 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Lus10035679 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Linaceae; Linum
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 735aa MW: 82623.5 Da PI: 6.6575 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 144.5 | 9.1e-45 | 59 | 205 | 2 | 134 |
DUF822 2 gsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpl..eeaeaagssasaspesslq.s 96
+g+++++kE+E++k+RER+RRai+++++aGLR++Gn++lp+raD+n+Vl+ L+reAGw+ve+DGttyr++s+p+ + ++ g +a ++ ++ +
Lus10035679 59 VKGKREREKEKERTKLRERHRRAITSRMLAGLRQYGNFPLPARADMNDVLASLAREAGWTVEADGTTYRQSSHPPplPHLATYGGGVRAVQSPTISaT 156
6899*********************************************************************9988444444444444333333335 PP
DUF822 97 slkssalaspve.sysaspksssfpsps.sldsislasa.........a 134
sl+++++++ ++ ++++ ++ + ++sp+ slds+ +++
Lus10035679 157 SLQACSVEAALDgQQHPALRMDGSLSPAaSLDSVVISDRdtrnegekfP 205
8888888777663778888888888885268888888775554444443 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 5.6E-46 | 60 | 214 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| SuperFamily | SSF51445 | 4.98E-152 | 234 | 675 | IPR017853 | Glycoside hydrolase superfamily |
| Gene3D | G3DSA:3.20.20.80 | 1.4E-167 | 237 | 669 | IPR013781 | Glycoside hydrolase, catalytic domain |
| Pfam | PF01373 | 8.4E-85 | 259 | 629 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 5.3E-55 | 274 | 288 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 5.3E-55 | 295 | 313 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 5.3E-55 | 317 | 338 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 5.3E-55 | 410 | 432 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 5.3E-55 | 483 | 502 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 5.3E-55 | 517 | 533 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 5.3E-55 | 534 | 545 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 5.3E-55 | 552 | 575 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 5.3E-55 | 588 | 610 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 735 aa Download sequence Send to blast |
MSTPIDDQNL LLDPYSQPLP TSSSNSHQHP QPQLRRPRGF AATAAAAASD ANGSNVSGVK 60 GKREREKEKE RTKLRERHRR AITSRMLAGL RQYGNFPLPA RADMNDVLAS LAREAGWTVE 120 ADGTTYRQSS HPPPLPHLAT YGGGVRAVQS PTISATSLQA CSVEAALDGQ QHPALRMDGS 180 LSPAASLDSV VISDRDTRNE GEKFPNSSPI NSVDCLDADQ LIQDVRSSEH VNDFTGSQYV 240 PVYVMLANGF INNFSQLIDS QGVRQELSRL KSLDVDGIVV DCWWGIVEAW SPQKYIWSGY 300 RELFHLVREF NLKLQVVMAF HEYGRNESSE VLISLPQWVL EIGKENRDIF FTDREGRRST 360 ECLSWGVDKE RVLKGRTGIE VYFDFMRSFR TEFDDLFVEG IITAVEISLG ASGELRYPSF 420 PERMGWRYPG IGEFQCYDKY LQRNLQRAAK LQELSVWAGS PDNAGQYNSR PHETGFFCER 480 GDYDSYFGRF FLRWYTQLLI SHADDVLSLA ALAFEDTKLI VKIPAVYWWY KTASHAAELT 540 AGFYNPSNQD GYSPVFKALK KHSVVVKLAC PGLQVSGHDE ALSDPEGLCW QVLNAAWDRG 600 LMVAGTNMLS LYGREGYMKV VDMAKPKNDP DHHHFSFFIY QQSSPLLQGM VCFPELDYFI 660 KCMHGCVLKV HKTLYNNFKE YKSVNDMRGC VGNMHHGLDL GVIPKLRRIC GLTGNLTLAQ 720 ELAEIILCVF SHEK* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wds_A | 1e-118 | 238 | 627 | 10 | 401 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
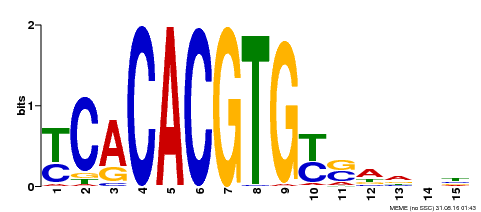
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Lus10035679 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021594907.1 | 0.0 | beta-amylase 8 isoform X3 | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | A0A2K2B4Z1 | 0.0 | A0A2K2B4Z1_POPTR; Beta-amylase | ||||
| STRING | Lus10035679 | 0.0 | (Linum usitatissimum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF11902 | 30 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.1 | 0.0 | beta-amylase 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Lus10035679 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



