 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Lus10036336 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Linaceae; Linum
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 364aa MW: 40003.6 Da PI: 4.9117 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 43.6 | 6.6e-14 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rgrWT eEd++l +++ G g+W+ ++ g++R++k+c++rw +yl
Lus10036336 14 RGRWTSEEDQILTKYILDNGEGSWRWLPKNAGLKRCGKSCRLRWINYL 61
8*********************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 49 | 1.4e-15 | 67 | 112 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg+ + eEd+ +v++ lG++ W++Ia++++ gRt++++k++w+++l
Lus10036336 67 RGNISDEEDDVIVKLQSSLGNR-WSLIASHLP-GRTDNEIKNYWNSHL 112
7899******************.*********.************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 14.632 | 9 | 61 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.87E-26 | 11 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.8E-8 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.6E-11 | 14 | 61 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.9E-19 | 15 | 68 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 9.38E-7 | 16 | 61 | No hit | No description |
| PROSITE profile | PS51294 | 24.598 | 62 | 116 | IPR017930 | Myb domain |
| SMART | SM00717 | 3.2E-15 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.2E-13 | 67 | 112 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 8.0E-24 | 69 | 116 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.79E-10 | 71 | 112 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009813 | Biological Process | flavonoid biosynthetic process | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 364 aa Download sequence Send to blast |
MGRAPCCEKI GLKRGRWTSE EDQILTKYIL DNGEGSWRWL PKNAGLKRCG KSCRLRWINY 60 LRGDLKRGNI SDEEDDVIVK LQSSLGNRWS LIASHLPGRT DNEIKNYWNS HLSRKVHSFR 120 RLVGNEPRLK LIVDTSSKVR TVGDGSNSKP LKSLSTKKGK TKKLVSNKGK IHNKVVVANS 180 GNDTLPSLEN VPPSDQNIRD RNAKEKQGVT LSATAPFSGI SDDGSIYDVE LDFLDGLMDS 240 EIANGPFALN EEGQSSVGHT ADTIMGQGHD VDDEGSGPIK SCDDVDTDGV VNIGSVEGTL 300 ECCWPTWSWE DVVVAGHEPL DDVVKFSNGD DDSLDLCKWS WDCEEVAGSS QEMDNDLVAW 360 LLS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1mse_C | 7e-23 | 12 | 116 | 2 | 105 | C-Myb DNA-Binding Domain |
| 1msf_C | 7e-23 | 12 | 116 | 2 | 105 | C-Myb DNA-Binding Domain |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00627 | PBM | Transfer from PK06182.1 | Download |
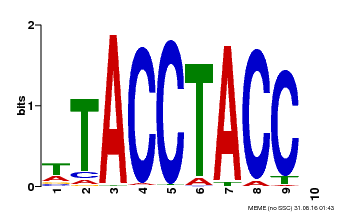
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Lus10036336 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| STRING | Lus10036336 | 0.0 | (Linum usitatissimum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF31 | 34 | 817 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47460.1 | 2e-70 | myb domain protein 12 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Lus10036336 |



