 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Lus10036567 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Linaceae; Linum
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 790aa MW: 85756.4 Da PI: 6.2499 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 66.2 | 4.4e-21 | 125 | 180 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++eLe+lF+++++p++++r eL+k+l L++rqVk+WFqNrR+++k
Lus10036567 125 KKRYHRHTPQQIQELEALFKECPHPDEKQRLELSKRLCLETRQVKFWFQNRRTQMK 180
688999***********************************************999 PP
| |||||||
| 2 | START | 163.7 | 1.4e-51 | 331 | 546 | 1 | 206 |
HHHHHHHHHHHHHHHHC-TT-EEEE....EXCCTTEEEEEEESSS......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EEEEEEEE CS
START 1 elaeeaaqelvkkalaeepgWvkss....esengdevlqkfeeskv.....dsgealrasgvvdmvlallveellddkeqWdetla....kaetlevi 85
ela++a++elvkka+ +ep+W ks e++n +e+l++f++ + + +ea+r++g+v+ ++ lve+l+d++ +W e+++ + +t++vi
Lus10036567 331 ELALAAMDELVKKAQTDEPLWLKSLdggrEMLNHEEYLRTFTPCIGlkpngFVTEASRETGMVIINSLALVETLMDSN-RWAEMFPcmiaRTSTTDVI 427
5899**************************************99999***9***************************.******************* PP
CTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--..-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE--SS CS
START 86 ssg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppe.sssvvRaellpSgiliepksnghskvtwvehvdlkgr 175
s+g g lqlm aelq+lsplvp R++ f+R+ vSvd ++++ + + +++lpSg+++++++ng+skvtwveh +++++
Lus10036567 428 SNGmggtrnGSLQLMHAELQVLSPLVPvREVNFLRFFVD----------VSVDAIREVSGgPPIFLSCRRLPSGCVVQDMPNGYSKVTWVEHGEYEES 515
***********************************9766..........66666666665557899******************************** PP
XXHHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 176 lphwllrslvksglaegaktwvatlqrqcek 206
++h+l+r l++sg+ +ga++wvatlqrqce+
Lus10036567 516 QIHQLYRLLIQSGMGFGAQRWVATLQRQCEC 546
*****************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.63E-20 | 110 | 182 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.6E-21 | 112 | 182 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.216 | 122 | 182 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 9.9E-18 | 123 | 186 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.23E-18 | 124 | 182 | No hit | No description |
| Pfam | PF00046 | 1.3E-18 | 125 | 180 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 157 | 180 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 32.322 | 322 | 549 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 1.26E-23 | 325 | 546 | No hit | No description |
| CDD | cd08875 | 1.24E-112 | 326 | 545 | No hit | No description |
| Pfam | PF01852 | 5.2E-44 | 331 | 546 | IPR002913 | START domain |
| SMART | SM00234 | 4.6E-34 | 331 | 546 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 5.49E-10 | 568 | 775 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009827 | Biological Process | plant-type cell wall modification | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0043481 | Biological Process | anthocyanin accumulation in tissues in response to UV light | ||||
| GO:0048765 | Biological Process | root hair cell differentiation | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 790 aa Download sequence Send to blast |
MSFGFLENGS PGGGRFVADY TTATTMSTTG GGMGSPPLLT PNFLSPNSFS KSMFSSPGLS 60 LALVRPQPNM DGVRQGDFGR ISENYETSVG RIRSREEEHD SRSGSDNLDG ASGDEQDAAD 120 KPPRKKRYHR HTPQQIQELE ALFKECPHPD EKQRLELSKR LCLETRQVKF WFQNRRTQMK 180 TQLERHENTL LRQENDKLRA ENMSIRDAMR NPMCSNCGGP AIIGDISLEE QHLRIENARL 240 KDELDRVCGL AGKFLGRPIS SLNNSLPPPM PNSSLELGVG NNDFGGFSTV AATLPVGPSD 300 HFGTPTANGL SIVTQNRPAS AGSLERSMLL ELALAAMDEL VKKAQTDEPL WLKSLDGGRE 360 MLNHEEYLRT FTPCIGLKPN GFVTEASRET GMVIINSLAL VETLMDSNRW AEMFPCMIAR 420 TSTTDVISNG MGGTRNGSLQ LMHAELQVLS PLVPVREVNF LRFFVDVSVD AIREVSGGPP 480 IFLSCRRLPS GCVVQDMPNG YSKVTWVEHG EYEESQIHQL YRLLIQSGMG FGAQRWVATL 540 QRQCECLAIL MSSTVPARDH TAAITASGRR SMLKLAQRMT DNFCAGVCAS TVHKWNKLNG 600 GNVEEDVRVM TRKSVDDPGE PPGIVLSAAT SVWLPVSPQF GCRCHPSGCL TFLGTKSMNA 660 NQSSMLILQE TCVDAAGSLV VYAPVDIPAM HVVMNGGDSA YVALLPSGFA IVPDGPGRRS 720 LGSTEDDAND NNGGGQTRAA ASGSLLTVAF QILVNSLPTA KLTVESVETV NNLISCTVQK 780 IKAALQCES* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of the tissue-specific accumulation of anthocyanins and in cellular organization of the primary root. {ECO:0000269|PubMed:10402424}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00421 | DAP | Transfer from AT4G00730 | Download |
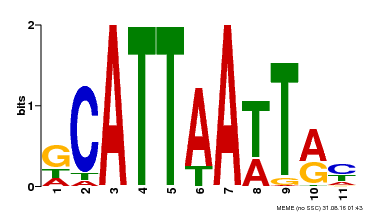
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Lus10036567 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002320755.1 | 0.0 | homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| Swissprot | Q0WV12 | 0.0 | ANL2_ARATH; Homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| TrEMBL | B9IC55 | 0.0 | B9IC55_POPTR; Uncharacterized protein | ||||
| STRING | Lus10036567 | 0.0 | (Linum usitatissimum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1192 | 34 | 106 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00730.1 | 0.0 | HD-ZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Lus10036567 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



