 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Lus10038022 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Linaceae; Linum
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 456aa MW: 48322.8 Da PI: 7.2824 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 53.1 | 7.2e-17 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g W++eEde+l++ ++++G g+W+++++ g+ R++k+c++rw +yl
Lus10038022 14 KGLWSPEEDEKLLRHITKFGHGCWSSVPKQAGLQRCGKSCRLRWINYL 61
678*******************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 44.7 | 3e-14 | 67 | 110 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
rg+++++E+ l++++++ lG++ W+ Ia+ ++ gRt++++k+ w++
Lus10038022 67 RGAFSQQEENLIIELHAVLGNR-WSQIAAQLP-GRTDNEIKNLWNS 110
89********************.*********.***********97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 1.7E-27 | 6 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 18.337 | 9 | 61 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 5.62E-29 | 12 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 9.4E-13 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 7.9E-15 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.48E-11 | 17 | 61 | No hit | No description |
| PROSITE profile | PS51294 | 25.261 | 62 | 116 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.3E-25 | 65 | 116 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 9.8E-13 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.3E-13 | 67 | 111 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 9.77E-9 | 69 | 109 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 456 aa Download sequence Send to blast |
MGRHSCCYKQ KLRKGLWSPE EDEKLLRHIT KFGHGCWSSV PKQAGLQRCG KSCRLRWINY 60 LRPDLKRGAF SQQEENLIIE LHAVLGNRWS QIAAQLPGRT DNEIKNLWNS CLKKKLRQRG 120 IDPVTHKPLS EVDIENKNIN NSSSSPGGAE GGGGGEENFV SSELNLLKQE KSNNMKALSS 180 VGGGGGLMGG GDVDKDFLLE RFAAVGSCHQ SPPPTDLVGH LNFPLHQLNY SAATSVADGR 240 LSTNSIPSLW YSGGGGGGGG KPFDMNSDFS SPTAAISIPS SFPYKPASVG GGGGGDNSXX 300 XXXXXXXXXX XXXXXXXXXX XSGAAAAGGG PPSSNNSSSG SSSSFFDTAA IFSWGIGDCG 360 TSEKGMGPAG AGATASAAQD DIKWPEYMNN PLLMSANQTQ LYSNNNDIKS ESHFLPESSS 420 SVWIHGGGDG GQQQFSDLSC PKDFQRLSAA YGHIN* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 4e-29 | 12 | 116 | 5 | 108 | B-MYB |
| 1h8a_C | 5e-29 | 11 | 116 | 24 | 128 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 111 | 117 | LKKKLRQ |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00427 | DAP | Transfer from AT4G01680 | Download |
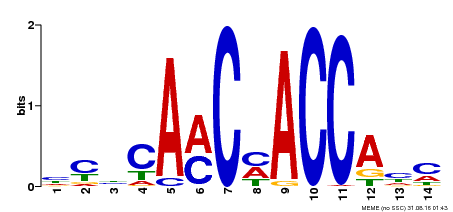
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Lus10038022 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021610810.1 | 1e-145 | transcription factor MYB61-like | ||||
| TrEMBL | A0A2C9WNK6 | 1e-144 | A0A2C9WNK6_MANES; Uncharacterized protein | ||||
| STRING | Lus10038022 | 0.0 | (Linum usitatissimum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1083 | 33 | 103 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G01680.3 | 6e-90 | myb domain protein 55 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Lus10038022 |



