 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Lus10041487 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Linaceae; Linum
|
||||||||
| Family | RAV | ||||||||
| Protein Properties | Length: 367aa MW: 40397.1 Da PI: 9.5959 | ||||||||
| Description | RAV family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 40.8 | 5.5e-13 | 64 | 112 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s+ykGV + +grW A+I++ ++r++lg+f ++eAa+ ++ a+++++g
Lus10041487 64 SKYKGVVPQP-NGRWGAQIYEK-----HQRVWLGTFNEEDEAARSYDIAALRFRG 112
89****9888.8*********3.....5**********99*************98 PP
| |||||||
| 2 | B3 | 97.2 | 1e-30 | 196 | 302 | 1 | 94 |
EEEE-..-HHHHTT-EE--HHH.HTT...........---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-.. CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh...........ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkld.. 85
f+k+ tpsdv+k++rlv+pk++ae+h +++++++ l +ed g++W+++++y+++s++yvltkGW++Fvk+++Lk+gD+v F+++
Lus10041487 196 FEKAVTPSDVGKLNRLVIPKQHAEKHfplqtggaaagVSSSTKGVLLNFEDAGGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKNLKAGDTVGFQRSvt 293
89****************************99999885555699**************************************************6532 PP
SSSEE..EE CS
B3 86 grsefelvv 94
g+++++l++
Lus10041487 294 GQDQKQLYI 302
335555555 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 4.48E-26 | 64 | 119 | No hit | No description |
| SuperFamily | SSF54171 | 1.31E-16 | 64 | 120 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 6.5E-8 | 64 | 112 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 20.587 | 65 | 120 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 3.2E-29 | 65 | 126 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 4.1E-20 | 65 | 120 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:2.40.330.10 | 5.5E-39 | 192 | 312 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 2.22E-29 | 195 | 291 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 7.73E-28 | 195 | 291 | No hit | No description |
| Pfam | PF02362 | 7.5E-28 | 196 | 304 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 13.916 | 196 | 308 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.1E-23 | 196 | 308 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009910 | Biological Process | negative regulation of flower development | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0048527 | Biological Process | lateral root development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 367 aa Download sequence Send to blast |
MDGISSSSAD ESTTTTTGES ISPLPTPPPA AKESLCRVGS GASSVVLDSD LSGIEAESRK 60 LPSSKYKGVV PQPNGRWGAQ IYEKHQRVWL GTFNEEDEAA RSYDIAALRF RGRDAVTNFK 120 KPSLSDASSD DESESESEFL NSHSKAEIVD MLRKHTYNDE LQQSRRRHHH HNRGKQSSTL 180 DSSDNIAARG AREQLFEKAV TPSDVGKLNR LVIPKQHAEK HFPLQTGGAA AGVSSSTKGV 240 LLNFEDAGGK VWRFRYSYWN SSQSYVLTKG WSRFVKEKNL KAGDTVGFQR SVTGQDQKQL 300 YINWKARSGE GVESNRLVCR VQPVQMVRLF GVNICKPVDG GCNGKRSMEE MVGFHCSKKQ 360 RVIGAL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wid_A | 2e-56 | 193 | 314 | 11 | 124 | DNA-binding protein RAV1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds specifically to bipartite recognition sequences composed of two unrelated motifs, 5'-CAACA-3' and 5'-CACCTG-3'. May function as negative regulator of plant growth and development. {ECO:0000269|PubMed:15040885, ECO:0000269|PubMed:9862967}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00139 | DAP | Transfer from AT1G13260 | Download |
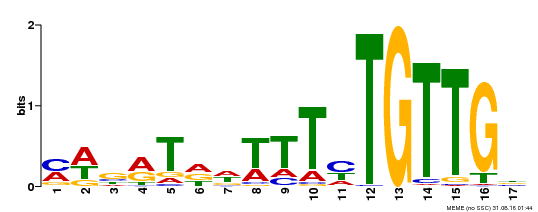
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Lus10041487 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated by brassinosteroid and zeatin. {ECO:0000269|PubMed:15040885}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021649928.1 | 1e-169 | AP2/ERF and B3 domain-containing transcription factor RAV1-like | ||||
| Swissprot | Q9ZWM9 | 1e-145 | RAV1_ARATH; AP2/ERF and B3 domain-containing transcription factor RAV1 | ||||
| TrEMBL | B9SEJ0 | 1e-167 | B9SEJ0_RICCO; DNA-binding protein RAV1, putative | ||||
| STRING | Lus10041487 | 0.0 | (Linum usitatissimum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2920 | 32 | 74 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G68840.2 | 1e-139 | related to ABI3/VP1 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Lus10041487 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



