 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MA_115846g0010 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Acrogymnospermae; Pinidae; Pinales; Pinaceae; Picea
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 1525aa MW: 168861 Da PI: 4.4062 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 42.4 | 1.6e-13 | 5 | 50 | 2 | 48 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
g W Ede+l+ av ++G+++W++I++ + ++++kqck rw+ +l
MA_115846g0010 5 GVWKNTEDEILKAAVMKYGKNQWARISSLLV-RKSAKQCKARWYEWL 50
78*****************************.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 32.1 | 2.7e-10 | 57 | 100 | 2 | 48 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
WT eEde+l+++ k++++ W+tIa +g Rt+ qc +r+ k+l
MA_115846g0010 57 TEWTREEDEKLLHLAKLMPTQ-WRTIAPIVG--RTPSQCLERYEKLL 100
68*****************99.********8..**********9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 24.223 | 1 | 54 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-17 | 2 | 54 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 7.0E-15 | 3 | 52 | IPR001005 | SANT/Myb domain |
| Pfam | PF13921 | 7.7E-13 | 7 | 67 | No hit | No description |
| CDD | cd00167 | 3.63E-12 | 7 | 50 | No hit | No description |
| SuperFamily | SSF46689 | 1.87E-20 | 30 | 104 | IPR009057 | Homeodomain-like |
| CDD | cd11659 | 5.29E-31 | 52 | 103 | No hit | No description |
| PROSITE profile | PS51294 | 16.443 | 55 | 104 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.7E-13 | 55 | 101 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.7E-12 | 55 | 102 | IPR001005 | SANT/Myb domain |
| Pfam | PF11831 | 2.0E-58 | 405 | 652 | IPR021786 | Pre-mRNA splicing factor component Cdc5p/Cef1 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009870 | Biological Process | defense response signaling pathway, resistance gene-dependent | ||||
| GO:0010204 | Biological Process | defense response signaling pathway, resistance gene-independent | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0009507 | Cellular Component | chloroplast | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1525 aa Download sequence Send to blast |
MIKGGVWKNT EDEILKAAVM KYGKNQWARI SSLLVRKSAK QCKARWYEWL DPSIKKTEWT 60 REEDEKLLHL AKLMPTQWRT IAPIVGRTPS QCLERYEKLL DAACVKDESY EPADDPRKLR 120 PGEIDPNPES KPARPDPVDM DEDEKEMLSE ARARLANTRG KKAKRKAREK QLEEARRLAS 180 LQKRRELKAA GIEGKQRRRK RKGIDYNAEI PFEKKPPAGF FDVSDEETRQ VEQPNFPTTI 240 EELEGKRRVD IEAQLRKQDV ARNKIMQRHD APSAVMQVNK LNDPEAVRKR AKLMLPSPQI 300 SDRELEEIAK MGYASDLVLG EDELAEGSGA ARALLANYGQ TPRLGMTPLR TPQRTPGGKG 360 DAIMMEAENQ ARLRESQTPL FGGENPELHP SDFSGVTPKK REIQTPNPIA TPMHTPGAVG 420 LTPRIGMTPS REFSMTPKGT PIRDELHINE GMETPDSSKA ESRRQAEIRR NLLAGLSDLP 480 VPKNEYQIVV PELPTEDIEV GYQMEEDMSD KLARERAEEA ARQAALLRKR SKVLQRDLPR 540 PPISSVDFIK NSLTRVDDDK GFVPTNFIEQ ADELIRKELV SLLEHDNAKY PFEEDSVKEK 600 KKGSKGAPNG RPQVPIPYID DFDVEELNEA AQLIEEEVSV LRVGMGHEDA SVDEFSEAHD 660 ACLEDMMYFP GRNSYGLGSV ASVTDKLAAT QNDFENVRNH MESETRKALR LEQKLKVLTQ 720 GYNMRAGKLW TQIEAAFKQA DTSGTELECF RSLHKQEQLA ASNRIEALRE AVNQQKEFER 780 QLQLRYQNLL VEQDNSQRHL QEQKEKMAHQ KEQENLQRLL KEQKEKMARQ KEQDNLERLL 840 QEQKEKLAHQ KDSRKENAID TQQSESAIAQ TDASHLENSS SEAQPFAISS KCNDVEMLDG 900 SGIAGEKTGE LDGSGIAEEK TERTGELDGS GIAEERTGEL DGSGIAEEKN GELDGSGIAE 960 EKNGELDGSG IAEEKNSELD GGSGIAEEKT GELDGSGIAE ERTGELDGSG IAEERTGELD 1020 GSGIAEEKNG ELDGSGIAEE KNGELDGSGI AEEKNSELDG GSGIAEEKTG ELDGSGIAKE 1080 KTDGSGIAEE KTGELDGSGI AEDKTSELDG SGIAEEKTSD NNSDMQIDCM PQSDGGHYNS 1140 DMQIDCVPQS DGHNNSDMQI DSVPQSDGHN PVLESEIVEQ NPHQTKDVMQ SDAKNIVVQD 1200 EKIVLQDENI DLITQSDGQN SVFRGDDVEQ VKHSTSLVAQ SDEQNDAIHN ESVEQVKNQQ 1260 ELADSSIYSQ NVINKDATTV SVEMQHPIQN STDGQNDAIH NENVLQLENQ QEAQALIYSQ 1320 NDINKDAAAV SVEMQPPIQN STDGQNDAIH NDNVLHVENQ QELAQPSIYS QNDINKDATG 1380 VSVAMQHPIQ NSTDGQNDAR YNENVGQVEN QQELAQSSTY SLIDINNDAA ALSVDMQQPI 1440 QNIDMVSSHG HLKEQLNDSD IQAQDVSAQT QQSVGDKTDT VMGVCKVTSQ EPVPLCTSDT 1500 EREIVVSTAQ VETAAAHTED VNLQS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5mqf_L | 0.0 | 1 | 795 | 5 | 797 | Cell division cycle 5-like protein |
| 5xjc_L | 0.0 | 1 | 795 | 5 | 797 | Cell division cycle 5-like protein |
| 5yzg_L | 0.0 | 1 | 795 | 5 | 797 | Cell division cycle 5-like protein |
| 5z56_L | 0.0 | 1 | 795 | 5 | 797 | Cell division cycle 5-like protein |
| 5z57_L | 0.0 | 1 | 795 | 5 | 797 | Cell division cycle 5-like protein |
| 5z58_L | 0.0 | 1 | 795 | 5 | 797 | Cell division cycle 5-like protein |
| 6ff4_L | 0.0 | 1 | 795 | 5 | 797 | Cell division cycle 5-like protein |
| 6ff7_L | 0.0 | 1 | 795 | 5 | 797 | Cell division cycle 5-like protein |
| 6icz_L | 0.0 | 1 | 795 | 5 | 797 | Cell division cycle 5-like protein |
| 6id0_L | 0.0 | 1 | 795 | 5 | 797 | Cell division cycle 5-like protein |
| 6id1_L | 0.0 | 1 | 795 | 5 | 797 | Cell division cycle 5-like protein |
| 6qdv_O | 0.0 | 1 | 795 | 5 | 797 | Cell division cycle 5-like protein |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 182 | 201 | KRRELKAAGIEGKQRRRKRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
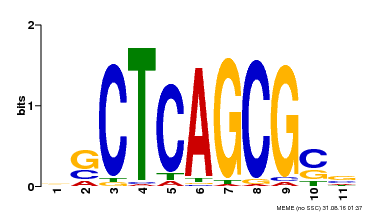
| UniProt | Component of the MAC complex that probably regulates defense responses through transcriptional control and thereby is essential for plant innate immunity. Possesses a sequence specific DNA sequence 'CTCAGCG' binding activity. Involved in mRNA splicing and cell cycle control. May also play a role in the response to DNA damage. {ECO:0000250|UniProtKB:Q99459, ECO:0000269|PubMed:17298883, ECO:0000269|PubMed:17575050, ECO:0000269|PubMed:8917598}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00028 | SELEX | Transfer from AT1G09770 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006857301.1 | 0.0 | cell division cycle 5-like protein | ||||
| Swissprot | P92948 | 0.0 | CDC5L_ARATH; Cell division cycle 5-like protein | ||||
| TrEMBL | U5DED6 | 0.0 | U5DED6_AMBTC; Uncharacterized protein | ||||
| STRING | ERN18768 | 0.0 | (Amborella trichopoda) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP3948 | 17 | 23 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09770.1 | 0.0 | cell division cycle 5 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



