 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MA_130615g0020 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Acrogymnospermae; Pinidae; Pinales; Pinaceae; Picea
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 313aa MW: 35658.7 Da PI: 4.5517 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 56.1 | 6.2e-18 | 45 | 99 | 2 | 56 |
T--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 2 rkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+k+++++ +q++ Le+ Fe ++++ e++ +LA++lgL+ rq+ vWFqNrRa++k
MA_130615g0020 45 KKKRRLSLQQVRSLEKTFEVENKLEPERKLQLAQELGLQPRQIAVWFQNRRARWK 99
677799************************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 123.2 | 1.3e-39 | 45 | 136 | 1 | 92 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreelk 92
+kkrrls +qv++LE++Fe e+kLeperK +la+eLglqprq+avWFqnrRAR+ktkqlE+dy +Lk +y+ lk++ e++++++++L++el+
MA_130615g0020 45 KKKRRLSLQQVRSLEKTFEVENKLEPERKLQLAQELGLQPRQIAVWFQNRRARWKTKQLERDYGQLKLNYECLKSNFEAVKHDNQKLTSELN 136
59**************************************************************************************9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 8.1E-20 | 27 | 102 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 3.14E-20 | 35 | 103 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 18.285 | 41 | 101 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 6.5E-17 | 44 | 105 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 2.6E-15 | 45 | 99 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.63E-16 | 45 | 102 | No hit | No description |
| PRINTS | PR00031 | 9.3E-6 | 72 | 81 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 76 | 99 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 9.3E-6 | 81 | 97 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 2.3E-14 | 101 | 141 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009788 | Biological Process | negative regulation of abscisic acid-activated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 313 aa Download sequence Send to blast |
MTKMETAEAL KFSIPSPLAG CPGSIHDDDE EEDEEECCAA GQQTKKKRRL SLQQVRSLEK 60 TFEVENKLEP ERKLQLAQEL GLQPRQIAVW FQNRRARWKT KQLERDYGQL KLNYECLKSN 120 FEAVKHDNQK LTSELNYLKE KWEETQKLQS DNTDLSPAKE GNNVSGMVED NPIPDTKSLD 180 RAMVCKQDGD AEIKVTSVAM IFNEKLERSE SESSAIVNEL ADNNNNNSPH SIDSHISTLS 240 LNCYYVPGPP VVSAGDEGLM EALCPRGLIR PKEEQINHHQ DFRADANFVQ EDPACNFFSV 300 DDTTVTLPWW EWP |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 44 | 48 | KKKRR |
| 2 | 44 | 49 | KKKRRL |
| 3 | 93 | 101 | RRARWKTKQ |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00271 | DAP | Transfer from AT2G22430 | Download |
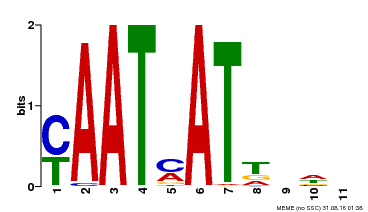
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT118795 | 0.0 | BT118795.1 Picea glauca clone GQ04010_F10 mRNA sequence. | |||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP129 | 16 | 189 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G22430.1 | 5e-42 | homeobox protein 6 | ||||



