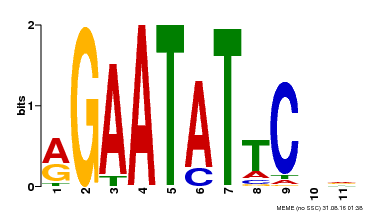
|
Plant Transcription
Factor Database
|
Transcription Factor Information
|
Basic
Information? help
Back to Top |
| TF ID |
MA_14087g0010 |
| Organism |
|
| Taxonomic ID |
|
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Acrogymnospermae; Pinidae; Pinales; Pinaceae; Picea
|
| Family |
G2-like |
| Protein Properties |
Length: 515aa MW: 57398.6 Da PI: 6.8811 |
| Description |
G2-like family protein |
| Gene Model |
| Gene Model ID |
Type |
Source |
Coding Sequence |
| MA_14087g0010 | genome | ConGenIE | View CDS |
|
| Signature Domain? help Back to Top |
 |
| No. |
Domain |
Score |
E-value |
Start |
End |
HMM Start |
HMM End |
| 1 | G2-like | 103.2 | 1.6e-32 | 260 | 315 | 1 | 56 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRla 56
k r++W+peLH+rFv+a++qLGGs++AtPk+i+elmkv+gLt+++vkSHLQkYRl+
MA_14087g0010 260 KSRRCWSPELHRRFVNALQQLGGSQVATPKQIRELMKVDGLTNDEVKSHLQKYRLH 315
68****************************************************97 PP
|
| Gene Ontology ? help Back to Top |
| GO Term |
GO Category |
GO Description |
| GO:0009266 | Biological Process | response to temperature stimulus |
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway |
| GO:0010452 | Biological Process | histone H3-K36 methylation |
| GO:0048579 | Biological Process | negative regulation of long-day photoperiodism, flowering |
| GO:1903507 | Biological Process | negative regulation of nucleic acid-templated transcription |
| GO:0005634 | Cellular Component | nucleus |
| GO:0043565 | Molecular Function | sequence-specific DNA binding |
| Sequence ? help Back to Top |
| Protein Sequence Length: 515 aa
Download sequence Send
to blast |
MRRHEEYIKG LEEERKKIEV FKRELPFCMQ LLNDAIEACK EQLADCERAS LHEDVQNAPN 60
TNGRPVLEEF IPIKKSYQAT ENKLEENHVA KKARFNNGDK PNWMISANLW NPDPETIDSR 120
KGGGISMEES LKEQDSQYQV HSFPSNHKLF SDSKHRMGGA FHPFSKERQV IAPRPVRSTV 180
ERTLPNLALS SAEREVDSSL VGNETVCLDA TTTTTARISR SKETTEVHTP VHEKEAVING 240
ANTSPTTSTP NTSQTQTQRK SRRCWSPELH RRFVNALQQL GGSQVATPKQ IRELMKVDGL 300
TNDEVKSHLQ KYRLHTRRPS PSPPSSNPQA PHLVVLGGIW VPPEYAAHAA AAAQQGQGGL 360
YNPLPTTVPC PPQSHYCQPS LPQGHEYYCQ MNPAASKLQL HHSVSHYCDQ QQEQQQQTPS 420
QSQNSPQGPL QCNGHSSGAR GTSADAGREE SVGEDAKSDS SSWKDEDSGE TTEVMNMGRG 480
LSLRKQPHEF LDEDDGDATD NRARVQMGKQ TLVGR
|
| 3D Structure ? help Back to Top |
 |
| PDB ID |
Evalue |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 6j4k_A | 1e-13 | 260 | 313 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 1e-13 | 260 | 313 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 9e-14 | 260 | 313 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 9e-14 | 260 | 313 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 9e-14 | 260 | 313 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 9e-14 | 260 | 313 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 1e-13 | 260 | 313 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 1e-13 | 260 | 313 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 1e-13 | 260 | 313 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 1e-13 | 260 | 313 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 1e-13 | 260 | 313 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 1e-13 | 260 | 313 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |