 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MA_18042g0010 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Acrogymnospermae; Pinidae; Pinales; Pinaceae; Picea
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 710aa MW: 75806.4 Da PI: 8.7817 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 35.4 | 1.9e-11 | 468 | 513 | 5 | 55 |
HHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 5 hnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
h+++Er RR+ri +++ L+el+P++ K +Ka++L + ++Y+k Lq
MA_18042g0010 468 HSIAERLRRERIAERMKALQELVPNS-----NKTDKASMLDEIIDYVKFLQ 513
99***********************7.....5******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 8.64E-17 | 461 | 522 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.453 | 463 | 512 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 3.9E-16 | 465 | 522 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 8.97E-13 | 467 | 517 | No hit | No description |
| Pfam | PF00010 | 8.6E-9 | 468 | 513 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 6.1E-14 | 469 | 518 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0048767 | Biological Process | root hair elongation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 710 aa Download sequence Send to blast |
MDVTAGVKID QELDPRSSFS FTPQGRRGKS TTLFQVSKFY KDDQKPAVRP ATEQFAGFNI 60 NGKHQCSAPI ISISAQAASE CKSFKFSKQA AGFESRDCEI IRRGRIPLRL DILASLVNFL 120 VDMQPSTSML GTGPVQGVSS VTACLSSSPQ VGLQEQMHSQ QHQINNQQQH FSSQFDQTNQ 180 PHQNMDDFLE QMLFMPQWSD VAGGKSPWEF NPNNAPQGNN ASNNNNSPQN LQSNTQKLFT 240 MSLMPPGIGS QGLNSQENDG HAGEHMQYSY DQSPFLANRL RQHQLSGNSS INQTPNQVSQ 300 GINETNTSPG RSMVLQLSTG NASASQLLAS MGNSPRGTAV MTRSPSTGGS CNGSDGGMLP 360 LPLSLGQAGK SGDMNEPGSR EEIEASFKSV NNARDSNLGG LFQPFAVSPR GVRPTGQNFH 420 AQPGQVPMQG YGGMPQPQHQ NPPPTGVGAA PPVRPRVRAR RGQATDPHSI AERLRRERIA 480 ERMKALQELV PNSNKTDKAS MLDEIIDYVK FLQLQVKVLS MSRLGGAGAV APLVADIPAE 540 GANAPQGTRT NGSQNSSPDG LALTERQVAK LMEEDMGTAM QYLQGKGLCL MPISLASAIN 600 NSGSRPQAPS TPSLQGLLAS NVNDRQVLEP SSNSALSALT STSMTIQPSI SSPGSGITEQ 660 GDNAHKAGNG TRNPKDSREA NSVTKSNGIG PALSKNAVKG EDEPQRRTGQ |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 472 | 477 | RLRRER |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00659 | PBM | Transfer from Pp3c14_15890 | Download |
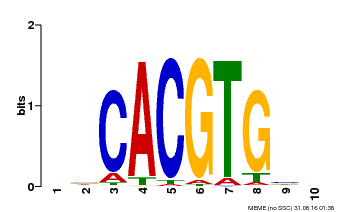
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT115043 | 0.0 | BT115043.1 Picea glauca clone GQ03607_N12 mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006857186.2 | 1e-101 | transcription factor bHLH66 | ||||
| Refseq | XP_011628047.1 | 1e-101 | transcription factor bHLH66 | ||||
| Refseq | XP_020530898.1 | 1e-101 | transcription factor bHLH66 | ||||
| Refseq | XP_020530899.1 | 1e-101 | transcription factor bHLH66 | ||||
| TrEMBL | U5DE19 | 1e-100 | U5DE19_AMBTC; Uncharacterized protein | ||||
| STRING | ERN18653 | 1e-101 | (Amborella trichopoda) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP58 | 16 | 313 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G24260.1 | 2e-45 | LJRHL1-like 1 | ||||



