 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MA_26114g0010 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Acrogymnospermae; Pinidae; Pinales; Pinaceae; Picea
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 333aa MW: 36624.1 Da PI: 7.8731 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 48.7 | 1.3e-15 | 160 | 206 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn E+rRR+riN+++ L+ l+P++ K +Ka++L +A+eY+k+Lq
MA_26114g0010 160 VHNLSEKRRRNRINEKMKALQNLIPNS-----NKTDKASMLDEAIEYLKKLQ 206
6*************************7.....5******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 4.71E-20 | 153 | 214 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 5.6E-20 | 153 | 215 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.885 | 156 | 205 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.23E-16 | 159 | 210 | No hit | No description |
| Pfam | PF00010 | 4.9E-13 | 160 | 206 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.4E-17 | 162 | 211 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 333 aa Download sequence Send to blast |
MAHKCLEPSF LVHNPVLWDD EDALNRNYNY NYYTDLWGGL PLSSSVATTA RTSAEDSRPE 60 VVAVSEALES DYGRLNNNNN NNVSADTQLS CETAATSSSG GSMSSPSNGN KVIKSSKKKV 120 LMGTKDIECQ SQKAQEDSGE NFKQCSTGTS SSKRSRAAEV HNLSEKRRRN RINEKMKALQ 180 NLIPNSNKTD KASMLDEAIE YLKKLQLQVQ MLSARSGIDI SSMRWLAQMP HLQIQQMPKA 240 CMTTDQHAGV SISMPVGSGL MNTNQGSEKR PLPLHDLYIS GALGNTAIPI NLPSARIDDH 300 QSCKMDRPQG HFTLPQLPTT TQEVHSALSL QEK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
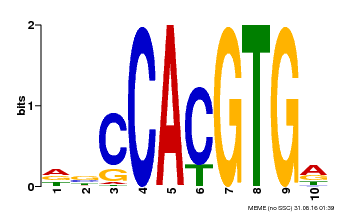
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EF084386 | 0.0 | EF084386.1 Picea sitchensis clone WS0272_M05 unknown mRNA. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001292767.1 | 2e-43 | transcription factor bHLH23 | ||||
| TrEMBL | A9NSU4 | 0.0 | A9NSU4_PICSI; Uncharacterized protein | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP258 | 16 | 128 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 4e-33 | phytochrome interacting factor 3 | ||||



