 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MA_8865g0010 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Acrogymnospermae; Pinidae; Pinales; Pinaceae; Picea
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 501aa MW: 55553.2 Da PI: 7.3675 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 100.2 | 1.3e-31 | 258 | 311 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
pr+rWt++LH++Fv+ave LGG+e+AtPk++lelm+vk+Ltl+hvkSHLQ+YR+
MA_8865g0010 258 PRMRWTSTLHAHFVHAVELLGGHERATPKSVLELMNVKDLTLAHVKSHLQMYRT 311
9****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 3.05E-15 | 255 | 312 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 5.8E-28 | 256 | 311 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.6E-23 | 258 | 311 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.0E-6 | 259 | 310 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009944 | Biological Process | polarity specification of adaxial/abaxial axis | ||||
| GO:0010158 | Biological Process | abaxial cell fate specification | ||||
| GO:0010229 | Biological Process | inflorescence development | ||||
| GO:0048481 | Biological Process | plant ovule development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 501 aa Download sequence Send to blast |
MLQEEIINDI SGPSGVITRC GGDLDLSLQI RPPNSKPSAT SSGGAEGRDL IGLDLWRRRN 60 TVKSNSTSLT ESSVITNLSS GSDRSQEDAT SLNFCFPNPG GLIDYNQLES ESRPTGLEDE 120 ARGLGFDHTT SARHSLSLVH GIGPIWINNP LKTENSGVVL VDKSLGFPGF REDSEPRIGD 180 LACRYIERLG RGIENQHREA LSRLLEADQN INRNSEIVSS ENCGLIPSSA VAPSPAEPNA 240 FRSRFMSKIP SKRSMRAPRM RWTSTLHAHF VHAVELLGGH ERATPKSVLE LMNVKDLTLA 300 HVKSHLQMYR TLKTTDKPAG PCDVFEVSSM QKSEDFMATY RVSSKQYNKS LSCNGEMDLG 360 PIQVTARQSA SPFNENQQKH QQNYYQNLQN NISSRSRNLF KDWQLPSSVN SLCQPRTRSS 420 IDQNEVELSG YQGNHHYIGR MEQNSLPQFP NSTSAIKPQS VSSQLINPKL ASFPDLDITL 480 GRPGWKLEHS DCPQELPLLK C |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 6e-17 | 259 | 313 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 6e-17 | 259 | 313 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 5e-17 | 259 | 313 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 5e-17 | 259 | 313 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 5e-17 | 259 | 313 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 5e-17 | 259 | 313 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 6e-17 | 259 | 313 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 6e-17 | 259 | 313 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 6e-17 | 259 | 313 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 6e-17 | 259 | 313 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 6e-17 | 259 | 313 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 6e-17 | 259 | 313 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00174 | DAP | Transfer from AT1G32240 | Download |
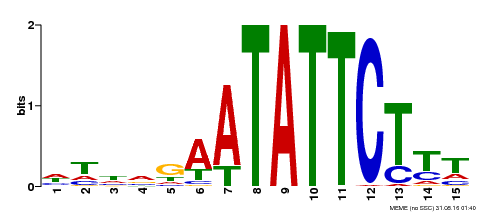
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT123184 | 0.0 | BT123184.1 Picea sitchensis clone WS0467_N16 unknown mRNA. | |||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP570 | 15 | 80 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G32240.1 | 7e-43 | G2-like family protein | ||||



