 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MA_937875g0010 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Acrogymnospermae; Pinidae; Pinales; Pinaceae; Picea
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 505aa MW: 57713.3 Da PI: 6.4073 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 102.8 | 2.1e-32 | 90 | 143 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
prlrWtp+LH++F++ave+LGG+++AtPk +l+lm+vkgLt++hvkSHLQ+YR+
MA_937875g0010 90 PRLRWTPDLHHCFMNAVERLGGQDRATPKLVLQLMDVKGLTIAHVKSHLQMYRS 143
8****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 10.653 | 86 | 146 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 7.1E-32 | 86 | 145 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 4.48E-15 | 88 | 146 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.5E-23 | 90 | 144 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 4.0E-8 | 91 | 142 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 505 aa Download sequence Send to blast |
MEVQGEQSSS NDGIELNSRK RSASSEDLEP PAKVFGGENT NPNTAKSSSN STVEGSEGKN 60 XXQSSSNSTV EGSEGKNGGT ARQYVRSKMP RLRWTPDLHH CFMNAVERLG GQDRATPKLV 120 LQLMDVKGLT IAHVKSHLQM YRSMKNDENG QGIGSQSEKH MEGRDHLPDC YSHSRTGLQQ 180 FENRRFLVND DYESAQYYNL FNRPTMQTFD HRLTTRYDRN PWAAHEDWLS RSYQIQLTNY 240 DNARGLYGWT SGNNLDWKNK REKALPVTRR DYKSELLGST IDPQLSVQWG KKQEEEKERT 300 CNLPEEDYFR PDFTGDLNTI SDQPTKHIRQ VEVDWSRWKH GREATVDSTN LGHGRIGTSC 360 IGLTDRNLQT APVKCNEPIT IDLEQESLPL QLQLQRELAQ QQLPRTLESP ESDYVRPERK 420 KKPKEPNLQL MLKHADDQDQ SWSEEEVDSS LSLSLFSKYQ EKETNSLKLA TSREGKPEPK 480 PFLQQANTDT HTPRGISTLD LTMSI |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 4e-17 | 91 | 145 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 4e-17 | 91 | 145 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 4e-17 | 91 | 145 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 4e-17 | 91 | 145 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 4e-17 | 91 | 145 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 4e-17 | 91 | 145 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 4e-17 | 91 | 145 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 4e-17 | 91 | 145 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 4e-17 | 91 | 145 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 4e-17 | 91 | 145 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 4e-17 | 91 | 145 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 4e-17 | 91 | 145 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00301 | DAP | Transfer from AT2G40260 | Download |
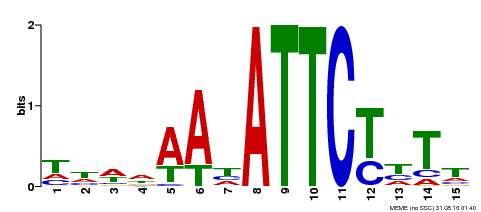
| |||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP285 | 15 | 123 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G40260.1 | 1e-35 | G2-like family protein | ||||



