 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MA_9412105g0010 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Acrogymnospermae; Pinidae; Pinales; Pinaceae; Picea
|
||||||||
| Family | LBD | ||||||||
| Protein Properties | Length: 197aa MW: 21934.6 Da PI: 8.9555 | ||||||||
| Description | LBD family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF260 | 133.6 | 7.6e-42 | 43 | 142 | 1 | 100 |
DUF260 1 aCaaCkvlrrkCakdCvlapyfpaeqpkkfanvhklFGasnvlkllkalpeeeredamsslvyeAearardPvyGavgvilklqqqleqlkael 94
+CaaCk+lrr+Ca++C+++pyf ++p+kfa vhk+FGasnv+k+l ++pe++r da++slvyeA++r+rdPvyG++g i++lqqq ++l++el
MA_9412105g0010 43 PCAACKLLRRRCAQECPFSPYFSPHEPQKFASVHKVFGASNVSKMLLEVPESQRADAANSLVYEANVRLRDPVYGCMGAISSLQQQAQSLQSEL 136
7********************************************************************************************* PP
DUF260 95 allkee 100
+++++e
MA_9412105g0010 137 NSVRAE 142
*99987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50891 | 25.416 | 42 | 143 | IPR004883 | Lateral organ boundaries, LOB |
| Pfam | PF03195 | 1.2E-41 | 43 | 140 | IPR004883 | Lateral organ boundaries, LOB |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016020 | Cellular Component | membrane | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 197 aa Download sequence Send to blast |
MSRERENFEN FSRRASFKQL ADADRMGRRQ ILGPPGTLNT ITPCAACKLL RRRCAQECPF 60 SPYFSPHEPQ KFASVHKVFG ASNVSKMLLE VPESQRADAA NSLVYEANVR LRDPVYGCMG 120 AISSLQQQAQ SLQSELNSVR AEIMRYKFRS EAAAMHHQPP GQSSHQASMM PSSSAVYSQG 180 SASTDYTAMN DHSNYYG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5ly0_A | 1e-45 | 35 | 146 | 3 | 114 | LOB family transfactor Ramosa2.1 |
| 5ly0_B | 1e-45 | 35 | 146 | 3 | 114 | LOB family transfactor Ramosa2.1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00283 | DAP | Transfer from AT2G30340 | Download |
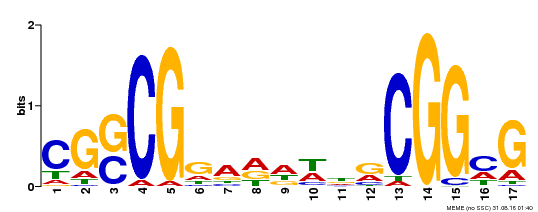
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT102233 | 0.0 | BT102233.1 Picea glauca clone GQ0171_H05 mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018821727.1 | 3e-90 | PREDICTED: LOB domain-containing protein 15-like | ||||
| Swissprot | Q8L5T5 | 1e-74 | LBD15_ARATH; LOB domain-containing protein 15 | ||||
| Swissprot | Q9AT61 | 3e-74 | LBD13_ARATH; LOB domain-containing protein 13 | ||||
| TrEMBL | A0A3S3NG72 | 5e-89 | A0A3S3NG72_9MAGN; LOB domain-containing protein 15 | ||||
| STRING | XP_007137607.1 | 4e-88 | (Phaseolus vulgaris) | ||||
| STRING | cassava4.1_031813m | 3e-88 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP60 | 16 | 318 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G40470.1 | 5e-68 | LOB domain-containing protein 15 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



