 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MDP0000147309 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Malus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 551aa MW: 59763 Da PI: 4.7636 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 53.8 | 4.3e-17 | 39 | 86 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT Ed +lv++v+++G g+W+++ ++ g+ R++k+c++rw ++l
MDP0000147309 39 KGPWTSAEDTILVEYVNRHGEGNWNAVQKHSGLFRCGKSCRLRWANHL 86
79******************************************9986 PP
| |||||||
| 2 | Myb_DNA-binding | 51.6 | 2.2e-16 | 92 | 135 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
+g++T++E+ l+v++++++G++ W++ a++++ gRt++++k++w++
MDP0000147309 92 KGAFTPDEERLIVELHAKMGNK-WARMAAHLP-GRTDNEIKNYWNT 135
799*******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 14.49 | 34 | 86 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 9.73E-30 | 38 | 133 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.8E-14 | 38 | 88 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.4E-15 | 39 | 86 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 6.1E-23 | 40 | 93 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 6.09E-11 | 41 | 86 | No hit | No description |
| PROSITE pattern | PS00175 | 0 | 54 | 63 | IPR001345 | Phosphoglycerate/bisphosphoglycerate mutase, active site |
| PROSITE profile | PS51294 | 26.12 | 87 | 141 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.3E-16 | 91 | 139 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.8E-15 | 92 | 135 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.7E-25 | 94 | 140 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 4.58E-12 | 94 | 135 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009789 | Biological Process | positive regulation of abscisic acid-activated signaling pathway | ||||
| GO:0043068 | Biological Process | positive regulation of programmed cell death | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0045926 | Biological Process | negative regulation of growth | ||||
| GO:0048235 | Biological Process | pollen sperm cell differentiation | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003824 | Molecular Function | catalytic activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 551 aa Download sequence Send to blast |
MSRTTNESED GMLYKDQIDS PLMDESNGGS GNGGIVLKKG PWTSAEDTIL VEYVNRHGEG 60 NWNAVQKHSG LFRCGKSCRL RWANHLRPNL KKGAFTPDEE RLIVELHAKM GNKWARMAAH 120 LPGRTDNEIK NYWNTRIKRR QRAGLPLYPP EVCLQALQES QQGQSSGGIN GADGAHNDSM 180 QANSFEIPDV VFDSLKGNNC VLPYVPELPD ISPGGMLMKG LGSSPYCGFM SPTMHRQKRL 240 RESTALFSTS GGSFKNGFPQ FDQFQNDTCD KIARTVGFPF PHDPDPTIPL SFGVIQGSHS 300 LSNGNSSASK PTTGAVKPEL PSLQYPETDL GSWSTSPPPP LLESVDAFIQ SPPXVGTFES 360 DCTSPRNSGL LDALLHEAKT LSSANNHSSE KSSNSSSVTP GDVAESSNLN ICETEWEEYR 420 DPISPLGHSA TSLFNECTPI SASGSSLDEP PPAETFTGCN VESELVDQAW TPDTERETAL 480 QLDYTRPDAI LASEWFGNSM HESIASLLGD DLAAECKHLA SGSPTSNQGL GFSSCPWNTM 540 LPFCEMSAQH P |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 3e-31 | 37 | 140 | 5 | 107 | B-MYB |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Mdo.16407 | 0.0 | fruit | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00490 | DAP | Transfer from AT5G06100 | Download |
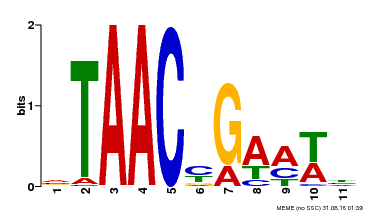
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008375384.2 | 0.0 | transcription factor GAMYB-like | ||||
| TrEMBL | A0A498JD71 | 0.0 | A0A498JD71_MALDO; Uncharacterized protein | ||||
| STRING | XP_008375383.1 | 0.0 | (Malus domestica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2047 | 34 | 85 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G06100.3 | 1e-122 | myb domain protein 33 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | MDP0000147309 |



