 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MDP0000193097 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Malus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 576aa MW: 63610.3 Da PI: 6.0295 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 53.1 | 7.4e-17 | 151 | 195 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r rWT+eE++++++a k++G W++I +++g ++t+ q++s+ qk+
MDP0000193097 151 RERWTEEEHKKFLEALKLYGRA-WRKIEKHVG-TKTAVQIRSHAQKF 195
78******************88.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.42E-16 | 145 | 200 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 21.095 | 146 | 200 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.0E-9 | 148 | 196 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.1E-16 | 149 | 198 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 9.4E-14 | 150 | 198 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.1E-14 | 151 | 194 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 5.53E-11 | 153 | 196 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 576 aa Download sequence Send to blast |
MIWKFSFSKL SKVNDDLETF GDRSELADGE RDVXKSTPIL EDRRAESKYG GVEIGTVIRK 60 WAGXRTEPCG EDRDKXLLWF LQAEGVRRNG VVTGRAEEEE QFGSTGSXGV VSAGNGISPS 120 AGGVQLKDDQ FSTGNDFTPK VRKPYTITKE RERWTEEEHK KFLEALKLYG RAWRKIEKHV 180 GTKTAVQIRS HAQKFFSKVA RYPNGGNATS EEPIDIPPPR PKKKPMRPYP RKLVHPLDKE 240 TSIVEQPTRS ASPNLSVSEQ ENQSPTSVLS VIGSDAMGST DSNTPSDGLS PDSSAAGVNS 300 ENLIHSEPNP LLEESGSPIX NCSLPNKQLT MKLELFPTDD VDASGGSAKE AFARSLKLFG 360 RTVLVTDSHR PSSRAAGTSQ SLPSDVQEEK PVQTSLPVNF TDMESASGNV EHVWDNFPYG 420 GHQGIYFMQF QNQNSDAVES GTNAYPXPWW TXCPPFIPFH KQQQQPVNEH VDSKKVDRED 480 SCTGSDAGSA SDEENRDKND ETQGDHYQQX SDKDREPNSV LQFKASANSA FSKIRASTGK 540 YRKGFVPYKR CTAERDAKSS TVASEDGDGK RICLSL |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Mdo.12650 | 0.0 | leaf | ||||
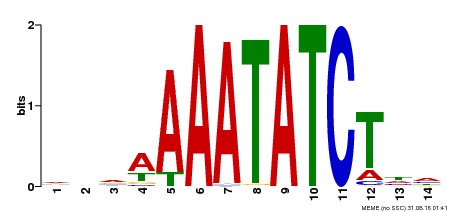
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00515 | DAP | Transfer from AT5G17300 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY826731 | 3e-35 | AY826731.1 Pisum sativum Myb2 mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009367121.1 | 0.0 | PREDICTED: protein REVEILLE 1-like isoform X1 | ||||
| TrEMBL | A0A498J5A4 | 0.0 | A0A498J5A4_MALDO; Uncharacterized protein | ||||
| STRING | XP_008381387.1 | 0.0 | (Malus domestica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2956 | 33 | 72 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G17300.1 | 1e-66 | MYB_related family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | MDP0000193097 |



