 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MDP0000197330 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Malus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 592aa MW: 64661.8 Da PI: 7.9132 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 61.8 | 1.4e-19 | 129 | 175 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd++++++v+++G+ W++Ia+ ++ gR +kqc++rw+++l
MDP0000197330 129 KGPWTQEEDDKIIELVARYGPTKWSLIAKSLP-GRIGKQCRERWHNHL 175
79******************************.*************97 PP
| |||||||
| 2 | Myb_DNA-binding | 55.1 | 1.7e-17 | 181 | 224 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
+ +WT eE++ l +a++++G++ W+ Ia+ ++ gRt++ +k++w++
MDP0000197330 181 KEAWTLEEELALMNAHQMHGNK-WAEIAKVLP-GRTDNAIKNHWNS 224
579*******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50090 | 7.991 | 56 | 123 | IPR017877 | Myb-like domain |
| SMART | SM00717 | 6.0E-6 | 60 | 125 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 4.74E-10 | 62 | 133 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 2.1E-16 | 63 | 131 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 3.34E-8 | 64 | 123 | No hit | No description |
| PROSITE profile | PS51294 | 32.788 | 124 | 179 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 7.49E-32 | 126 | 222 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.3E-18 | 128 | 177 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.4E-19 | 129 | 175 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.60E-16 | 131 | 175 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 2.5E-28 | 132 | 178 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 6.4E-24 | 179 | 230 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 22.577 | 180 | 230 | IPR017930 | Myb domain |
| SMART | SM00717 | 2.7E-16 | 180 | 228 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.5E-16 | 181 | 224 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.46E-12 | 183 | 226 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 592 aa Download sequence Send to blast |
MAGVKLEECC LENKQSTAAS SSSVSQGSGS GSAIIKSPGA CSPASASPTQ RRTSGPIRRA 60 KGGWTPQEDE TLRVAVAAFK GKSWKKIGGN DSAQQMCLTN LLNAEFFPDR SEVQCLHRWQ 120 KVLNPELVKG PWTQEEDDKI IELVARYGPT KWSLIAKSLP GRIGKQCRER WHNHLNPDIK 180 KEAWTLEEEL ALMNAHQMHG NKWAEIAKVL PGRTDNAIKN HWNSSLKKKL EFYLATGKLP 240 PPPKYGTKET RTSATKKLLV CSTKGSDSTA QTSSGNADMS KLDEDGKDQF ESAAPPQDMG 300 ASPSVHPNES ADSEGVKCNL RLSFMDLSCS NSDSGPKFEN FRVNRMPQIE NSVINSQAES 360 ENCVFNNQLD EDRVKTTSLP VETPGYGSLY YEPPPPESCA QFDSGFSNMH XLQYDYTSSP 420 NLSPISFFTP PSANGSGLCN QSPESILKIA AKTFPNTPSI LRKRKTIGQE QLPRNKVAIV 480 DXESGIVDCE SVREQERHKN SAEVSGSQDG SSCESPTNNG ISTIGSNGKA FNASPPYRLR 540 ARRTAVFKSV EKQLEFTCDK EYDANSKSME LSANGGSAVI KDSTHPTKMG VT |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 5e-65 | 60 | 230 | 5 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 5e-65 | 60 | 230 | 5 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Mdo.15998 | 0.0 | bud| fruit | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed both in proliferating and maturing stages of leaves. {ECO:0000269|PubMed:26069325}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds 5'-AACGG-3' motifs in gene promoters (By similarity). Transcription repressor that regulates organ growth. Binds to the promoters of G2/M-specific genes and to E2F target genes to prevent their expression in post-mitotic cells and to restrict the time window of their expression in proliferating cells (PubMed:26069325). {ECO:0000250|UniProtKB:Q94FL9, ECO:0000269|PubMed:26069325}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00482 | DAP | Transfer from AT5G02320 | Download |
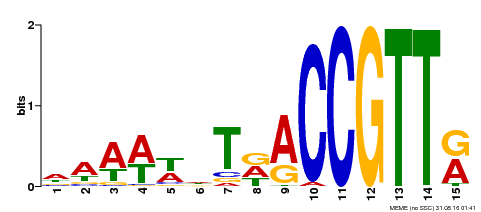
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Slightly induced by ethylene and salicylic acid (SA). {ECO:0000269|PubMed:16463103}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | DQ074460 | 0.0 | DQ074460.1 Malus x domestica MYB4 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008369900.2 | 0.0 | transcription factor MYB3R-5 isoform X1 | ||||
| Swissprot | Q6R032 | 1e-161 | MB3R5_ARATH; Transcription factor MYB3R-5 | ||||
| TrEMBL | A0A498K7G0 | 0.0 | A0A498K7G0_MALDO; Uncharacterized protein | ||||
| STRING | XP_008369900.1 | 0.0 | (Malus domestica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF6616 | 34 | 49 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G02320.2 | 1e-150 | myb domain protein 3r-5 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | MDP0000197330 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



