 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MDP0000216714 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Malus
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 176aa MW: 20725.7 Da PI: 10.0153 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 164 | 5.2e-51 | 26 | 152 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevlsk. 95
+ pG+rFhPtdeelv +yL++kve+k+++l + ik+vdiyk++PwdLpk ++++kewyfF++r +ky+++ r+nr+tksg+Wkatg dk+v+s
MDP0000216714 26 MLPGYRFHPTDEELVRFYLRRKVENKPIRL-DLIKHVDIYKYDPWDLPK--ASGDKEWYFFCRRGRKYRNSIRPNRVTKSGFWKATGIDKPVYSVg 118
579***************************.99***************4..35899**************************************85 PP
NAM 96 .kgelvglkktLvfykgrapkgektdWvmheyrl 128
++ +glkk+Lv+y+g+a kg+ktdW+mhe+rl
MDP0000216714 119 dFHNCIGLKKSLVYYRGSAGKGTKTDWMMHEFRL 152
55566***************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 5.49E-54 | 22 | 165 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 50.222 | 26 | 176 | IPR003441 | NAC domain |
| Pfam | PF02365 | 1.0E-26 | 28 | 152 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005992 | Biological Process | trehalose biosynthetic process | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0006561 | Biological Process | proline biosynthetic process | ||||
| GO:0009718 | Biological Process | anthocyanin-containing compound biosynthetic process | ||||
| GO:0010120 | Biological Process | camalexin biosynthetic process | ||||
| GO:0042538 | Biological Process | hyperosmotic salinity response | ||||
| GO:1900056 | Biological Process | negative regulation of leaf senescence | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 176 aa Download sequence Send to blast |
MEEVDQKMMS ASDSNNMIYK DQDANMLPGY RFHPTDEELV RFYLRRKVEN KPIRLDLIKH 60 VDIYKYDPWD LPKASGDKEW YFFCRRGRKY RNSIRPNRVT KSGFWKATGI DKPVYSVGDF 120 HNCIGLKKSL VYYRGSAGKG TKTDWMMHEF RLPASSNTDT SIKNSTQEAV RLIHLK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3ulx_A | 6e-46 | 21 | 152 | 10 | 140 | Stress-induced transcription factor NAC1 |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, root caps, cotyledons, tips and margin of young leaves, senescent regions of fully expanded leaves and floral tissues, including old sepals, petals, staments, mature anthers and pollen grains. Not detected in the abscission zone of open flowers, emerging lateral roots and root meristematic zones. {ECO:0000269|PubMed:22345491}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
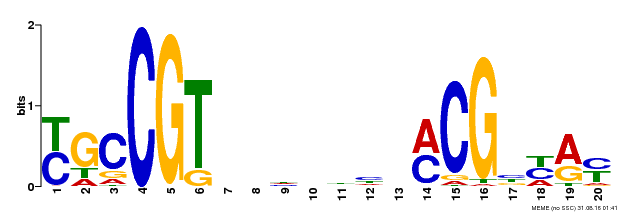
| UniProt | Transcription factor that binds to the 5'- RRYGCCGT-3' consensus core sequence. Central longevity regulator. Negative regulator of leaf senescence. Modulates cellular H(2)O(2) levels and enhances tolerance to various abiotic stresses through the regulation of DREB2A. {ECO:0000269|PubMed:22345491}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00310 | DAP | Transfer from AT2G43000 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by H(2)O(2), paraquat, ozone, 3-aminotriazole and salt stress. {ECO:0000269|PubMed:22345491}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EU794447 | 3e-32 | EU794447.1 Malus floribunda clone M18-6Bs Vf apple scab resistance protein HcrVf2-like gene, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008359791.2 | 1e-125 | transcription factor JUNGBRUNNEN 1-like | ||||
| Swissprot | Q9SK55 | 7e-76 | NAC42_ARATH; Transcription factor JUNGBRUNNEN 1 | ||||
| TrEMBL | A0A498K0B6 | 1e-123 | A0A498K0B6_MALDO; Uncharacterized protein | ||||
| STRING | XP_008350676.1 | 1e-125 | (Malus domestica) | ||||
| STRING | XP_008359791.1 | 1e-123 | (Malus domestica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1013 | 34 | 112 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G43000.1 | 3e-78 | NAC domain containing protein 42 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | MDP0000216714 |



