 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MDP0000234807 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Malus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 321aa MW: 35405.7 Da PI: 10.5639 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 36 | 1.6e-11 | 22 | 72 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHT.....TTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmg.....kgRtlkqcksrwqk 46
+++WT eE+e l+ +v ++G g W+tI R+ ++k++w++
MDP0000234807 22 KQKWTSEEEEALKAGVLKHGAGKWRTILTDPEfntilHLRSNVDLKDKWRN 72
79***********************************88**********99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 11.221 | 17 | 78 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 5.46E-15 | 19 | 84 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 7.5E-13 | 21 | 73 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 7.8E-6 | 21 | 76 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.3E-7 | 22 | 72 | IPR001005 | SANT/Myb domain |
| CDD | cd11660 | 6.42E-17 | 23 | 74 | No hit | No description |
| SMART | SM00526 | 5.4E-15 | 137 | 204 | IPR005818 | Linker histone H1/H5, domain H15 |
| PROSITE profile | PS51504 | 24.951 | 138 | 209 | IPR005818 | Linker histone H1/H5, domain H15 |
| SuperFamily | SSF46785 | 5.99E-17 | 139 | 214 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Gene3D | G3DSA:1.10.10.10 | 3.0E-15 | 143 | 213 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Pfam | PF00538 | 2.9E-10 | 146 | 202 | IPR005818 | Linker histone H1/H5, domain H15 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006334 | Biological Process | nucleosome assembly | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0000786 | Cellular Component | nucleosome | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003691 | Molecular Function | double-stranded telomeric DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0043047 | Molecular Function | single-stranded telomeric DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 321 aa Download sequence Send to blast |
MKLSWQISFW KGSGFRLMGA PKQKWTSEEE EALKAGVLKH GAGKWRTILT DPEFNTILHL 60 RSNVDLKDKW RNINVTAIWG SRQKAKLALK RNLPTPKHEN NNNPLAVSTI IQSHKEVVDA 120 KPLAISGGKS QTTESKDSKH PISRLDHLIL EAITNLKEPG GSDRAAIVMY IEEQYWAPPN 180 LKKLLSSKLK HMTTNRKLVK VKHRYRIPAT SEKRRSSSAL LPDGKQKDSS RTDKSDVNIL 240 TKSQVDADLT KMRSMTAQEA AAAAAQAVAE AEAAIAAAEE AAREAEAAEA EAEAAQVFAK 300 AAMKALKCRK LGKVLLSNFT S |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Mdo.12025 | 0.0 | bud| fruit| leaf | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Ubiquitous. {ECO:0000269|PubMed:15060584}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds preferentially double-stranded telomeric repeats, but it can also bind to the single G-rich telomeric strand. | |||||
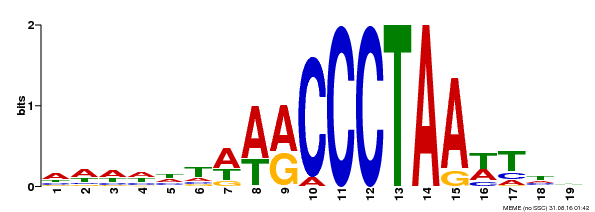
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00597 | DAP | Transfer from AT5G67580 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | DQ074477 | 0.0 | DQ074477.1 Malus x domestica MYBR6 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_028963643.1 | 0.0 | telomere repeat-binding factor 2 isoform X1 | ||||
| Refseq | XP_028963644.1 | 0.0 | telomere repeat-binding factor 2 isoform X1 | ||||
| Swissprot | Q9FJW5 | 7e-80 | TRB2_ARATH; Telomere repeat-binding factor 2 | ||||
| TrEMBL | A0A498KH78 | 0.0 | A0A498KH78_MALDO; Uncharacterized protein | ||||
| STRING | XP_008341201.1 | 0.0 | (Malus domestica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1453 | 33 | 101 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G49950.3 | 1e-73 | telomere repeat binding factor 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | MDP0000234807 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



