 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MDP0000251332 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Malus
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 387aa MW: 40625.3 Da PI: 6.059 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 76.4 | 3.6e-24 | 294 | 356 | 1 | 63 |
XXXXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 1 ekelkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklksev 63
e+elkr++ kq+NRe+ArrsR+RK+ae+eeL+ +v++L++eN+ L++el +l++e++kl+se+
MDP0000251332 294 ERELKRQKXKQSNRESARRSRLRKQAECEELQARVEVLSNENHGLREELHRLSEECEKLTSEN 356
89***********************************************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF07777 | 5.2E-31 | 46 | 133 | IPR012900 | G-box binding protein, multifunctional mosaic region |
| Pfam | PF16596 | 2.1E-10 | 155 | 266 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 5.0E-20 | 288 | 353 | No hit | No description |
| Pfam | PF00170 | 1.3E-21 | 294 | 356 | IPR004827 | Basic-leucine zipper domain |
| SMART | SM00338 | 6.0E-21 | 294 | 358 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 12.633 | 296 | 359 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 3.52E-11 | 297 | 353 | No hit | No description |
| CDD | cd14702 | 4.17E-24 | 299 | 349 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 301 | 316 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010310 | Biological Process | regulation of hydrogen peroxide metabolic process | ||||
| GO:0010629 | Biological Process | negative regulation of gene expression | ||||
| GO:0090342 | Biological Process | regulation of cell aging | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 387 aa Download sequence Send to blast |
MALHYPSQVS GNTWGSTVPF SLVAPIDQFV KGHGQPSLFG DNWEEGTPPK PSKQASTAQE 60 IPTPPSYPDW SSSMQAYYGP GGTPPPFFAS TVASPTPHPY MWGAQHPMMP PYGTPVPYPA 120 MYPPGGVYAH PSMVTTPGAP QPAPELEGKG SDGKERASTK KTKGTAGNAS LAGGKAVESG 180 KATSGSGNDG ASQSGESGSE GSSDGSDDNA NHQEYGTNKK GSFDKMLADG ANAQNSTGAI 240 QASVPGKPVS MPGTNLNIGM DLWNASPAGA GAAKVRGNPS GAPSAGGEHW IQDERELKRQ 300 KXKQSNRESA RRSRLRKQAE CEELQARVEV LSNENHGLRE ELHRLSEECE KLTSENTNIK 360 EELTRVCGPD LVANLEQQPG GGEGKNS |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 310 | 316 | RRSRLRK |
| 2 | 310 | 317 | RRSRLRKQ |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Mdo.26786 | 0.0 | bud| leaf| stem | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Found in both light and dark grown leaves. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds to the G-box motif (5'-CCACGTGG-3') of the rbcS-1A gene promoter (PubMed:1373374). G-box and G-box-like motifs are cis-acting elements defined in promoters of certain plant genes which are regulated by such diverse stimuli as light-induction or hormone control. Binds to the G-box motif 5'-CACGTG-3' of LHCB2.4 (At3g27690) promoter. May act as transcriptional activator in light-regulated expression of LHCB2.4. Probably binds DNA as monomer. DNA-binding activity is redox-dependent (PubMed:22718771). {ECO:0000269|PubMed:1373374, ECO:0000269|PubMed:22718771}. | |||||
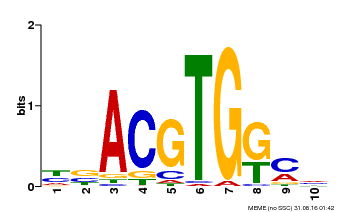
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00039 | PBM | Transfer from AT4G36730 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001315805.1 | 0.0 | G-box-binding factor 1-like | ||||
| Swissprot | P42774 | 1e-120 | GBF1_ARATH; G-box-binding factor 1 | ||||
| TrEMBL | A0A498KG46 | 0.0 | A0A498KG46_MALDO; Uncharacterized protein | ||||
| TrEMBL | D9ZIR0 | 0.0 | D9ZIR0_MALDO; BZIP domain class transcription factor | ||||
| STRING | XP_008363114.1 | 0.0 | (Malus domestica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF5933 | 34 | 50 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G36730.1 | 1e-107 | G-box binding factor 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | MDP0000251332 |



