 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MDP0000265114 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Malus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 327aa MW: 37122.4 Da PI: 8.3781 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 56.3 | 7.5e-18 | 60 | 105 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
rg W + Ede+l ++v+++G+++W++Ia++++ gR++k+c++rw++
MDP0000265114 60 RGHWRPAEDEKLRELVERYGPHNWNAIAEKLQ-GRSGKSCRLRWFNQ 105
899*****************************.***********996 PP
| |||||||
| 2 | Myb_DNA-binding | 53.6 | 5.3e-17 | 112 | 155 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
r+++T+eE+e+l+ ++ +G++ W+ Iar ++ gRt++ +k++w+
MDP0000265114 112 RNPFTEEEEERLLVSHRIHGNR-WAVIARLFP-GRTDNAVKNHWHV 155
789*******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 17.203 | 55 | 106 | IPR017930 | Myb domain |
| SMART | SM00717 | 2.9E-15 | 59 | 108 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 3.56E-29 | 60 | 153 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 1.4E-17 | 60 | 105 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.3E-27 | 61 | 113 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 3.31E-14 | 63 | 104 | No hit | No description |
| PROSITE profile | PS51294 | 23.321 | 107 | 161 | IPR017930 | Myb domain |
| SMART | SM00717 | 6.3E-15 | 111 | 159 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.0E-14 | 112 | 154 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.41E-11 | 114 | 154 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 2.5E-20 | 114 | 160 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:2000652 | Biological Process | regulation of secondary cell wall biogenesis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 327 aa Download sequence Send to blast |
MVSDSPRAVG VVSQDVASIA GVSSTIIIVS VVIALPLRST SSSSSSSSSS SLLSAIMCTR 60 GHWRPAEDEK LRELVERYGP HNWNAIAEKL QGRSGKSCRL RWFNQLDPRI NRNPFTEEEE 120 ERLLVSHRIH GNRWAVIARL FPGRTDNAVK NHWHVIMARR CRERSRLNHS KLIRNTSSIT 180 AQTSFMSIIT NNNEPKSSSK QDHPLQINCD IGTWNLDSYN YIGKYYSCSD RNYHQYRPAF 240 THNPPPFPKE FYPNTCSVTM HQDHNKHQPI EFYDFLQVNT ESNGSEVIDN ARKDEEEVDQ 300 EGMGQQKSTV TGLPFINFLA VNNGTSS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1mse_C | 3e-32 | 57 | 160 | 1 | 104 | C-Myb DNA-Binding Domain |
| 1msf_C | 3e-32 | 57 | 160 | 1 | 104 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in stamen (PubMed:19325888). Present in roots and siliques, and, at low levels, in leaves and flowers (PubMed:21399993). Expressed in stems, especially in fibers and, at lower levels, in xylems (PubMed:18952777, PubMed:21399993). {ECO:0000269|PubMed:18952777, ECO:0000269|PubMed:19325888, ECO:0000269|PubMed:21399993}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that confers sensitivity to abscisic acid (ABA) and salt, but tolerance to drought (PubMed:21399993). Regulates secondary cell wall (SCW) biosynthesis, especially in interfascicular and xylary fibers (PubMed:18952777, PubMed:23781226). {ECO:0000269|PubMed:18952777, ECO:0000269|PubMed:21399993, ECO:0000269|PubMed:23781226}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00145 | DAP | Transfer from AT1G17950 | Download |
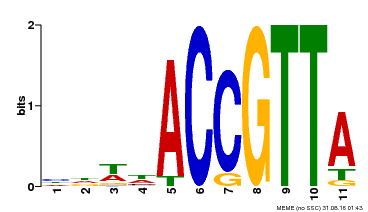
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By abscisic acid (PubMed:16463103, PubMed:21399993). Accumulates in response to salt (PubMed:21399993). Triggered by MYB46 and MYB83 in the regulation of secondary cell wall biosynthesis (PubMed:19674407, PubMed:22197883). {ECO:0000269|PubMed:16463103, ECO:0000269|PubMed:19674407, ECO:0000269|PubMed:21399993, ECO:0000269|PubMed:22197883}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008393477.1 | 0.0 | transcription factor MYB54 | ||||
| Refseq | XP_017192802.1 | 0.0 | transcription factor MYB54 | ||||
| Swissprot | Q6R0C4 | 7e-86 | MYB52_ARATH; Transcription factor MYB52 | ||||
| TrEMBL | A0A498I0N6 | 0.0 | A0A498I0N6_MALDO; Uncharacterized protein | ||||
| STRING | XP_008393477.1 | 0.0 | (Malus domestica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF230 | 34 | 243 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G17950.1 | 2e-87 | myb domain protein 52 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | MDP0000265114 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



