 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MDP0000290409 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Malus
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1153aa MW: 129397 Da PI: 5.9463 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 179.5 | 3.9e-56 | 65 | 181 | 2 | 118 |
CG-1 2 lkekkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqrr 97
l+ k+rwl++ ei++iL+n++k+++++ ++++p+ gsl+L++rk++ryfrkDG++w+kkkdgktv+E+he+LK g+v+vl+cyYah+een++fqrr
MDP0000290409 65 LEAKHRWLRPAEICEILQNYQKFQIASVPANKPPGGSLFLFDRKVLRYFRKDGHNWRKKKDGKTVKEAHERLKAGSVDVLHCYYAHGEENENFQRR 160
6679******************************************************************************************** PP
CG-1 98 cywlLeeelekivlvhylevk 118
+yw+Lee+l++ivlvhy+evk
MDP0000290409 161 SYWMLEEALQHIVLVHYREVK 181
******************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 82.292 | 60 | 186 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.8E-78 | 63 | 181 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 1.3E-49 | 66 | 179 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 1.7E-5 | 565 | 651 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 6.47E-18 | 566 | 651 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF01833 | 1.9E-7 | 566 | 650 | IPR002909 | IPT domain |
| Gene3D | G3DSA:1.25.40.20 | 5.0E-16 | 748 | 861 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 16.839 | 749 | 860 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 5.00E-12 | 749 | 858 | No hit | No description |
| SuperFamily | SSF48403 | 1.76E-15 | 751 | 861 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 0.01 | 799 | 828 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 9.618 | 799 | 831 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 1000 | 838 | 867 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 1.17E-7 | 969 | 1024 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 14 | 973 | 995 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.364 | 975 | 1003 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.053 | 976 | 994 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.021 | 996 | 1018 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.432 | 997 | 1021 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0013 | 999 | 1018 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1153 aa Download sequence Send to blast |
MWLTKLLVVG PRQSSSHAFT ISLYAQTSCV RGLSXSESLT THIEFMEETK RYGLGNQLDI 60 AQILLEAKHR WLRPAEICEI LQNYQKFQIA SVPANKPPGG SLFLFDRKVL RYFRKDGHNW 120 RKKKDGKTVK EAHERLKAGS VDVLHCYYAH GEENENFQRR SYWMLEEALQ HIVLVHYREV 180 KGNRTNYNHI QGTEEGVPYS HETEEVALNS EMDNSVSSSF NPSSFQMRSQ ATDATSLSSA 240 QASEFEDAES VYNHQASSQL QPFLELLQPK AEKTNAGVST AFYPMSFSNE YQEKLSAIPG 300 VNFSSHTQAY RKEDVKDAGV TYDPRKNLNS TLWDGALGNF TTGFQPLPFQ PXISATHSDS 360 TGIISKQENE TFGHLFTNNF GKKQMYEDRP RVQQGWQTLE ANSSGSSSWP VDQNLHSNTA 420 YDVSTRLYEG VHASNLLNSL VCHXDSDKTN DYSMPNDLQI QPSNPEQEYH LKSISKRNET 480 IEGSYKHAFA TKPLLDEGLK KLDSFNRWMS KELGDVDETQ TQSNSETYWD TVESENGVDE 540 SSVPLQVRLD SYMLGPSLSQ DQLFSIIDFS PNWAYENSEI KVLITGRFLK SQEAESCKWS 600 CMFGEVEVPA EVIADGVLRC YTPIHKAGRI PFYVTCSNRL ACSEIREFEY RVGQIPDYDA 660 KDDYTGCTNE ILNMRFGKLL SLSSSSPTFD PTSIAENSEL ISKIDLLLKN DNGEWDRMLQ 720 LTSDEDFSLE RVEDQLLQQL LKEKLRAWLL QKLAAGGKGP SVLDEGGQGV LHFGAALGYD 780 WVLLPTITAG VSVNFRDVDG WTALHWAAFH GRERTVASLI SLGAAPGLLT DPRTKYPAGR 840 TPADLASAQG HKGIAGYLAE STLSDHLSFL NLDIKEGNNA EISGAKAVET VSEQIATPIG 900 NGDLTGGLSL RDSLTAVCNA TQAAARIHQV LRVKSFQRKQ LKEFGSDNFG ISDEDALSLI 960 AVKSHKPGKR DEHVDAAAIR IQNKFRSWKG RKDYLIIRQR IVKIQAHVRG HQVRKXYRKI 1020 VWSVGIVEKI ILRWRRKGSG LRGFKPEALA EPPSMQASSS KDDDYDVLKE GRKQTEQRLQ 1080 KALARVKSMI QYPEARDQYS RLLNVVTEIQ ETKVVYDSSM NSSDGRADMD DDLVDIAALL 1140 DEDDVCMPTA APE |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2cxk_A | 1e-14 | 566 | 653 | 9 | 91 | calmodulin binding transcription activator 1 |
| 2cxk_B | 1e-14 | 566 | 653 | 9 | 91 | calmodulin binding transcription activator 1 |
| 2cxk_C | 1e-14 | 566 | 653 | 9 | 91 | calmodulin binding transcription activator 1 |
| 2cxk_D | 1e-14 | 566 | 653 | 9 | 91 | calmodulin binding transcription activator 1 |
| 2cxk_E | 1e-14 | 566 | 653 | 9 | 91 | calmodulin binding transcription activator 1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Mdo.2496 | 1e-171 | bud| fruit| leaf | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, stems, leaves, carpels, and siliques, but not in stigmas or other parts of the flower. {ECO:0000269|PubMed:11162426, ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:14581622}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3'. Binds calmodulin in a calcium-dependent manner in vitro (PubMed:12218065). Regulates transcriptional activity in response to calcium signals (Probable). Involved in freezing tolerance in association with CAMTA1 and CAMTA2 (PubMed:23581962). Required for the cold-induced expression of DREB1B/CBF1, DREB1C/CBF2, ZAT12 and GOLS3 (PubMed:19270186). Involved in response to cold. Contributes together with CAMTA5 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). Involved together with CAMTA2 and CAMTA4 in the positive regulation of a general stress response (GSR) (PubMed:25039701). Involved in the regulation of GSR amplitude downstream of MEKK1 (PubMed:25157030). Involved in the regulation of a set of genes involved in defense responses against pathogens (PubMed:18298954). Involved in the regulation of both basal resistance and systemic acquired resistance (SAR) (PubMed:21900483). Acts as negative regulator of plant immunity (PubMed:19122675, PubMed:21900483, PubMed:22345509, PubMed:28407487). Binds to the promoter of the defense-related gene EDS1 and represses its expression (PubMed:19122675). Binds to the promoter of the defense-related gene NDR1 and represses its expression (PubMed:22345509). Involved in defense against insects (PubMed:23072934, PubMed:22371088). Required for tolerance to the generalist herbivore Trichoplusia ni, and contributes to the positive regulation of genes associated with glucosinolate metabolism (PubMed:23072934). Required for tolerance to Bradysia impatiens larvae. Mediates herbivore-induced wound response (PubMed:22371088). Required for wound-induced jasmonate accumulation (PubMed:23072934, PubMed:22371088). Involved in the regulation of ethylene-induced senescence by binding to the promoter of the senescence-inducer gene EIN3 and repressing its expression (PubMed:22345509). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:18298954, ECO:0000269|PubMed:19122675, ECO:0000269|PubMed:19270186, ECO:0000269|PubMed:21900483, ECO:0000269|PubMed:22345509, ECO:0000269|PubMed:22371088, ECO:0000269|PubMed:23072934, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25157030, ECO:0000269|PubMed:28351986, ECO:0000269|PubMed:28407487, ECO:0000305|PubMed:11925432}. | |||||
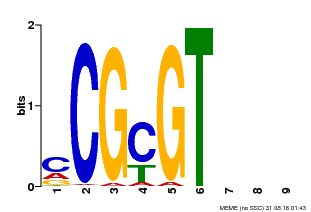
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00042 | PBM | Transfer from AT2G22300 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, salt, wounding, ethylene and methyl jasmonate (PubMed:11162426, PubMed:12218065). Induced by infection with the fungal pathogen Golovinomyces cichoracearum (powdery mildew) and the bacterial pathogen Pseudomonas syringae pv tomato strain DC3000 (PubMed:22345509). {ECO:0000269|PubMed:11162426, ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:22345509}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008338581.2 | 0.0 | calmodulin-binding transcription activator 3 isoform X1 | ||||
| Swissprot | Q8GSA7 | 0.0 | CMTA3_ARATH; Calmodulin-binding transcription activator 3 | ||||
| TrEMBL | A0A498HVW2 | 0.0 | A0A498HVW2_MALDO; Uncharacterized protein | ||||
| STRING | XP_008338581.1 | 0.0 | (Malus domestica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF8284 | 27 | 35 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G22300.2 | 0.0 | signal responsive 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | MDP0000290409 |



