|
Plant Transcription
Factor Database
|
Transcription Factor Information
|
Basic
Information? help
Back to Top |
| TF ID |
MDP0000320521 |
| Organism |
|
| Taxonomic ID |
|
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Malus
|
| Family |
BBR-BPC |
| Protein Properties |
Length: 313aa MW: 34463.5 Da PI: 9.7373 |
| Description |
BBR-BPC family protein |
| Gene Model |
| Gene Model ID |
Type |
Source |
Coding Sequence |
| MDP0000320521 | genome | GDR | View CDS |
|
| Signature Domain? help Back to Top |
 |
| No. |
Domain |
Score |
E-value |
Start |
End |
HMM Start |
HMM End |
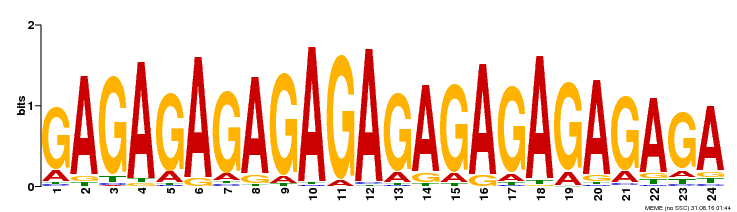
| 1 | GAGA_bind | 340.5 | 4.3e-104 | 35 | 313 | 1 | 301 |
GAGA_bind 1 mdddgsrernkgyyepaaslkenlglqlmssiaerdaki....rernlalsekkaavaerd.........maflqrdkalaernkalverdnklla 83
mddd+ + rn+gyyep+ +k +l+lqlmss+aerd+k+ r+ + + + ++a+++rd m++ +r++++++ ++k+l+
MDP0000320521 35 MDDDALNMRNWGYYEPS--FKGHLSLQLMSSMAERDTKPfvpgRDPTV-MVSANGAYHPRDcvvsdaqlpMNY-MRESWVNQ--------RDKFLN 118
9***9999*******99..**********************9999999.999*********999999999999.88899998........7899** PP
GAGA_bind 84 lllvenslasalpvgvqvlsgtksidslqqlsepqledsavelreeeklealpieeaaeeakekkkkkkrqrakkpkekkakkkkkksekskkkvk 179
+++ a+p++ vl++t+ ++slq l+++ r+e+ + iee + + ke+ ++kkrq + pk++k kk++k k+++
MDP0000320521 119 MMP-------ANPNYGAVLPETSGAHSLQI-----LQPPEPS-RDER---VGRIEE-PVVPKEVGTSKKRQGGGAPKAPKVKKPRKA-----KDST 192
999.......4688999*********9999.....4444333.3333...334433.566778888888999999999999999982.....3344 PP
GAGA_bind 180 kesaderskaekksidlvlngvslDestlPvPvCsCtGalrqCYkWGnGGWqSaCCtttiSvyPLPvstkrrgaRiagrKmSqgafkklLekLaae 275
++s+ +r+k kks+d+v+ng+++D+s++P+P+CsCtGa++qCY+WG+GGWqSaCCtt++S+yPLP+stkrrgaRiagrKmSqgafkk+LekLaae
MDP0000320521 193 NPSV-PRVKPAKKSLDVVINGINMDISGIPIPICSCTGAPQQCYRWGCGGWQSACCTTNVSMYPLPMSTKRRGARIAGRKMSQGAFKKVLEKLAAE 287
4554.79***************************************************************************************** PP
GAGA_bind 276 GydlsnpvDLkdhWAkHGtnkfvtir 301
Gy+++np+DL+ hWAkHGtnkfvt+r
MDP0000320521 288 GYNFANPIDLRFHWAKHGTNKFVTLR 313
************************97 PP
|
| Gene Ontology ? help Back to Top |
| GO Term |
GO Category |
GO Description |
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated |
| GO:0009723 | Biological Process | response to ethylene |
| GO:0050793 | Biological Process | regulation of developmental process |
| GO:0005634 | Cellular Component | nucleus |
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding |
| GO:0043565 | Molecular Function | sequence-specific DNA binding |
| Sequence ? help Back to Top |
| Protein Sequence Length: 313 aa
Download sequence Send
to blast |
MASEGYPMVI FIVVQGLSLE GLNDVCLSAD SVAXMDDDAL NMRNWGYYEP SFKGHLSLQL 60
MSSMAERDTK PFVPGRDPTV MVSANGAYHP RDCVVSDAQL PMNYMRESWV NQRDKFLNMM 120
PANPNYGAVL PETSGAHSLQ ILQPPEPSRD ERVGRIEEPV VPKEVGTSKK RQGGGAPKAP 180
KVKKPRKAKD STNPSVPRVK PAKKSLDVVI NGINMDISGI PIPICSCTGA PQQCYRWGCG 240
GWQSACCTTN VSMYPLPMST KRRGARIAGR KMSQGAFKKV LEKLAAEGYN FANPIDLRFH 300
WAKHGTNKFV TLR
|
| Expression --
Description ? help
Back to Top |
| Source |
Description |
| Uniprot | DEVELOPMENTAL STAGE: Early detected in the floral meristem and floral organ primordia. At later stages, expressed in all floral organs and in particular in the ovule. {ECO:0000269|PubMed:15722463}. |
| Uniprot | TISSUE SPECIFICITY: Expressed in seedlings, leaves and pistils. Detected in the base of flowers and tips of carpels, in leaf and sepal vasculature, in young rosette, in the lateral and tip of primary roots, and in the whole ovule. {ECO:0000269|PubMed:14731261, ECO:0000269|PubMed:21435046}. |
| Functional Description ? help
Back to Top |
| Source |
Description |
| UniProt | Transcriptional regulator that specifically binds to GA-rich elements (GAGA-repeats) present in regulatory sequences of genes involved in developmental processes. Negatively regulates the homeotic gene AGL11/STK, which controls ovule primordium identity, by a cooperative binding to purine-rich elements present in the regulatory sequence leading to DNA conformational changes. {ECO:0000269|PubMed:14731261, ECO:0000269|PubMed:15722463}. |
| Annotation --
Nucleotide ? help
Back to Top |
| Source |
Hit ID |
E-value |
Description |
| GenBank | KM244747 | 0.0 | KM244747.1 Malus hupehensis basic pentacysteine 1 protein (BPC1) mRNA, complete cds. |
| GenBank | KP657685 | 0.0 | KP657685.1 Malus baccata basic pentacysteine 1 protein (BPC1) mRNA, complete cds. |