 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MDP0000334047 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Malus
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 308aa MW: 35617.5 Da PI: 6.7925 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 162.4 | 1.7e-50 | 15 | 139 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevlskk 96
lppGfrFhPtdeelv++y ++k+++++l + +i+e+d+yk++Pw+Lp+++ +ekewyfFs+rd+ky++g+r+nra+ +gyWkatg dk++ +
MDP0000334047 15 LPPGFRFHPTDEELVNHYXCRKCASQPLAV-PIIREIDLYKFDPWQLPEMALYGEKEWYFFSPRDRKYPNGSRPNRAAGTGYWKATGADKHIGK-- 107
79*************************999.88***************7666789*************************************98.. PP
NAM 97 gelvglkktLvfykgrapkgektdWvmheyrl 128
++ g+kk Lvfy g+apkg kt+W+mheyrl
MDP0000334047 108 PKALGIKKALVFYAGKAPKGIKTNWIMHEYRL 139
778***************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 2.35E-61 | 10 | 168 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 59.063 | 15 | 168 | IPR003441 | NAC domain |
| Pfam | PF02365 | 3.9E-26 | 16 | 139 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009788 | Biological Process | negative regulation of abscisic acid-activated signaling pathway | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 308 aa Download sequence Send to blast |
MSGGGDEQQQ QQLELPPGFR FHPTDEELVN HYXCRKCASQ PLAVPIIREI DLYKFDPWQL 60 PEMALYGEKE WYFFSPRDRK YPNGSRPNRA AGTGYWKATG ADKHIGKPKA LGIKKALVFY 120 AGKAPKGIKT NWIMHEYRLA NVDRSAAAAK KNQNLRLDDW VLCRIYNKKG SIEKYNVTTK 180 MTKYPEIXDE QKPDMTIMPP PPHAASNTHH MMDSSMDSVP RLPQTTTDYS SCSEHVLSPE 240 VTWEKEVQSE LKWSNQLENS FNTLDNQFLN YMDGFSDIMD PFGGQLQMEQ QQPYQLQDMY 300 SYLHTQFX |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3ulx_A | 2e-81 | 6 | 174 | 6 | 174 | Stress-induced transcription factor NAC1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Mdo.24695 | 0.0 | bud| leaf | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00121 | DAP | Transfer from AT1G01720 | Download |
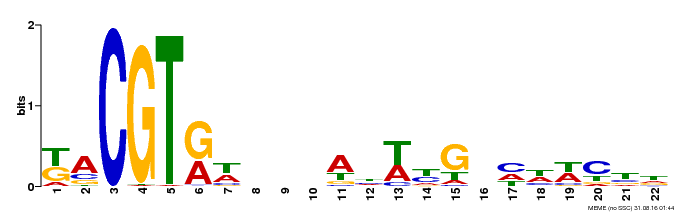
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KF198129 | 0.0 | KF198129.1 Malus hupehensis NAC domain protein (NAC143) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008394193.2 | 0.0 | NAC domain-containing protein 2 | ||||
| Swissprot | Q39013 | 1e-118 | NAC2_ARATH; NAC domain-containing protein 2 | ||||
| TrEMBL | A0A498HLG8 | 0.0 | A0A498HLG8_MALDO; Uncharacterized protein | ||||
| STRING | XP_008394193.1 | 0.0 | (Malus domestica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1258 | 34 | 108 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G01720.1 | 1e-114 | NAC family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | MDP0000334047 |



