 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MDP0000439540 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Malus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 450aa MW: 48033.5 Da PI: 8.0134 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 48.9 | 1.2e-15 | 272 | 317 | 5 | 55 |
HHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 5 hnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn+ Er+RRd+iN+++ +L++l+P++ K +Ka++L +++eY+k+Lq
MDP0000439540 272 HNQSERKRRDKINQRMKTLQKLVPNS-----SKTDKASMLDEVIEYLKQLQ 317
9************************8.....7******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 1.27E-18 | 265 | 328 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.815 | 267 | 316 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 1.2E-17 | 272 | 324 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.45E-17 | 272 | 321 | No hit | No description |
| Pfam | PF00010 | 3.0E-13 | 272 | 317 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 2.2E-18 | 273 | 322 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009567 | Biological Process | double fertilization forming a zygote and endosperm | ||||
| GO:0009506 | Cellular Component | plasmodesma | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 450 aa Download sequence Send to blast |
MSQRVPSWDL DKTTTTPPAP RLSSRSHSLH TAPTDVPLLD YEVAELTWEN GEVAMHGLGP 60 PRLLAKPLST TKYTTWDKPR ASGTLESIVN QATSTALPPP LKPPFDSSSG GGSGAKELVT 120 WFDRHRAVAT PSTTVTMDAL VPCRNQSDDP SSQMIESISV PGVISGTGMV GCSTRVDSCS 180 GATGAATHDD DTLLSGKGTS LARVPETPEW SSRSQSVSGS ATFGIDSQHV TLDSTKASDY 240 DSGEVGDGDD RKKRSTGKSS ISTKRSRAAA IHNQSERKRR DKINQRMKTL QKLVPNSSKT 300 DKASMLDEVI EYLKQLQAQV QMISRMNMPT IMLPMAMQQL QMSMMAAAPR NMGMGMGMGM 360 GMGMDMNNIG RPSIPGIPPI LHPSAFMPMA ASWDGTGGDR LPAASTGVMP DPMSAFLACQ 420 SQPMTMDAYS MMAAMYQQFH QPPASSSKG* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 275 | 280 | ERKRRD |
| 2 | 276 | 281 | RKRRDK |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Mdo.15898 | 0.0 | bud| leaf | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in stems and flowers. {ECO:0000269|PubMed:12679534}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Required during the fertilization of ovules by pollen. {ECO:0000269|PubMed:15634699}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00034 | PBM | Transfer from AT4G00050 | Download |
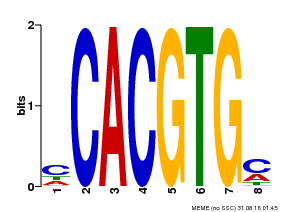
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By cold treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KP689423 | 1e-113 | KP689423.1 Prunus pseudocerasus isolate Unigene11342 bHLH transcription factor mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008375803.1 | 0.0 | transcription factor UNE10 isoform X1 | ||||
| Swissprot | Q8GZ38 | 9e-93 | UNE10_ARATH; Transcription factor UNE10 | ||||
| TrEMBL | A0A498JF35 | 0.0 | A0A498JF35_MALDO; Uncharacterized protein | ||||
| STRING | XP_008375803.1 | 0.0 | (Malus domestica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF5676 | 33 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00050.1 | 4e-93 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | MDP0000439540 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



