 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MDP0000607330 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Malus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 501aa MW: 54903.5 Da PI: 5.4059 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 51.7 | 2e-16 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+grWT+eEde+l+++++ G g+W++ ++ g+ R++k+c++rw +yl
MDP0000607330 14 KGRWTAEEDEILLNYIQANGEGSWRSLPKNAGLLRCGKSCRLRWINYL 61
79******************************99************97 PP
| |||||||
| 2 | Myb_DNA-binding | 48.3 | 2.3e-15 | 67 | 112 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg+ + +E+++++++++ lG++ W++Ia+ ++ gRt++++k++w+++l
MDP0000607330 67 RGNISSQEEDIIIKLHASLGNR-WSLIASQLP-GRTDNEIKNYWNSHL 112
78999*****************.*********.************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 6.5E-22 | 5 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 16.625 | 9 | 61 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.3E-27 | 11 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 3.0E-12 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.8E-14 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 8.91E-10 | 16 | 61 | No hit | No description |
| PROSITE profile | PS51294 | 24.507 | 62 | 116 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.7E-24 | 65 | 116 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 4.4E-15 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.2E-13 | 67 | 112 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.21E-10 | 71 | 112 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009813 | Biological Process | flavonoid biosynthetic process | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 501 aa Download sequence Send to blast |
MGRAPCCEKV GLKKGRWTAE EDEILLNYIQ ANGEGSWRSL PKNAGLLRCG KSCRLRWINY 60 LRADLKRGNI SSQEEDIIIK LHASLGNRWS LIASQLPGRT DNEIKNYWNS HLSRKIGTFR 120 RPATTTVITT EISTSXPPAG DEVSAAMELG PPKRRGGRTS RWXMKKNKTY STTKPKGLKA 180 RLRQSHHKHD NIAAAAADDD RLNNAHEAIA LPTKSNNKNV DTMQDGYVLL MEVPDQQQET 240 RGGGIAMPAT VIDHQKETDG EKLILGPHPH GHDDDDMNEY VGINGGLLGF TDYLMDDINE 300 DEILDPNGVM ALSSSEINIH QDADHFPDAV ISDHQDTELP SSCDQLVICP NKVMTTTTTT 360 TASSDDHHDQ ELESSSCGCN LSSSSSISNS NGKVVTGDLH SCGSSLSLTN TSNCDTEDGH 420 TAGGFGNHDQ ILWDWESVLS NAGTGHDYLW DGHDRQNMLS WLREGESATA AGDDDHADAM 480 DFXXXXPPXX KSSGYQARKH * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h8a_C | 4e-23 | 12 | 116 | 25 | 128 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Mdo.14481 | 0.0 | bud | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00627 | PBM | Transfer from PK06182.1 | Download |
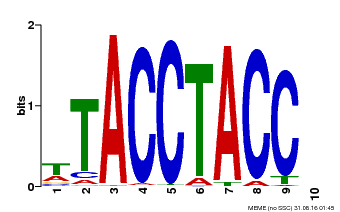
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | DQ074470 | 0.0 | DQ074470.1 Malus x domestica MYB22 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008369060.2 | 0.0 | LOW QUALITY PROTEIN: transcription factor MYB11 | ||||
| TrEMBL | A0A498K4R8 | 0.0 | A0A498K4R8_MALDO; Uncharacterized protein | ||||
| STRING | XP_008369060.1 | 0.0 | (Malus domestica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF31 | 34 | 817 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47460.1 | 1e-71 | myb domain protein 12 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | MDP0000607330 |



