 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MDP0000795432 | ||||||||
| Common Name | MYBR8 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Malus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 462aa MW: 50257.5 Da PI: 7.3187 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 52.7 | 9.9e-17 | 54 | 98 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r +WT+eE++++++a k++G g W+ I +++g ++t+ q++s+ qk+
MDP0000795432 54 RAKWTEEEHQKFLEALKLYGRG-WRQIEEHVG-TKTAVQIRSHAQKF 98
569*******************.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 8.97E-17 | 48 | 104 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 21.604 | 49 | 103 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.1E-9 | 51 | 101 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 4.2E-16 | 52 | 101 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 4.8E-13 | 53 | 101 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.3E-14 | 54 | 97 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.33E-11 | 56 | 99 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 462 aa Download sequence Send to blast |
MAVEDKIEAT GSNASVAVGN CSSNGDAQSN AEICSFGSDH APKVRKPYTI TKQRAKWTEE 60 EHQKFLEALK LYGRGWRQIE EHVGTKTAVQ IRSHAQKFFS KVSKESCGPS EGSIRPIEIP 120 PPRPKRKPVH PYPRKSVDCL NGTPERSPSP QFSAQGKDQQ SPPSVLSAQG SDLLGSAALD 180 QHNRSSTPTS CTTDMHSTGP SHFEKENDSM TFSSSAEEVQ GSSPSVQLSA DSVLEKFPSV 240 NYRSGSKHTV STEEDAGNSG ASTSIKLFGR TVSVLDSEKQ SPTGAKDSKT PTSKVEQDNC 300 ERENDELVQT PRSDLLDTSL TLGGIVGNLY PSACGGTTTW EHQKHSPNPE APLPWWDMYH 360 GLPCFHIRSC NQNSAQIQIR VDSCADETIK QTERSFGGAV ESLGEKHVES VSSLREETRS 420 APCSSRKGFV PYKRCLADVN KNSSAIVSEN MRDRRRARVC S* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Mdo.13219 | 0.0 | bud| fruit| leaf | ||||
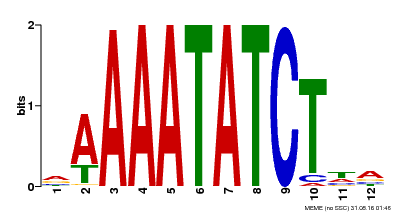
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00146 | DAP | Transfer from AT1G18330 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KM001671 | 0.0 | KM001671.1 Pyrus x bretschneideri transcription factor MYBR (MYBR) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008393486.2 | 0.0 | protein REVEILLE 7-like | ||||
| TrEMBL | D9ZJ85 | 0.0 | D9ZJ85_MALDO; MYBR domain class transcription factor | ||||
| STRING | XP_009355963.1 | 0.0 | (Pyrus x bretschneideri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF7313 | 32 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G18330.1 | 9e-51 | MYB_related family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | MDP0000795432 |



