 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MDP0000875621 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Malus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 322aa MW: 35277.6 Da PI: 9.6438 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 40.9 | 4.8e-13 | 132 | 176 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT+eE+ ++ + ++lG+g+W+ I+r ++Rt+ q+ s+ qky
MDP0000875621 132 PWTEEEHRTFLIGLEKLGKGDWRGISRNYVTTRTPTQVASHAQKY 176
8*******************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50158 | 8.532 | 3 | 18 | IPR001878 | Zinc finger, CCHC-type |
| PROSITE profile | PS51294 | 19.284 | 125 | 181 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.2E-17 | 126 | 182 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 3.8E-18 | 128 | 180 | IPR006447 | Myb domain, plants |
| Gene3D | G3DSA:1.10.10.60 | 7.1E-11 | 129 | 175 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.8E-9 | 129 | 179 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.1E-10 | 132 | 176 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.48E-8 | 132 | 177 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 322 aa Download sequence Send to blast |
MGRKCSHCGN IGHNSRTCAN FRGTTSSTSF VGFRLFGVQL HHDHDHHIMM SSSNMNNNSA 60 NYHVSMPMKK SFSMDCLPTT TSSASSSSPS NSSLSPSRLS MDENICASDK ASIGYLSDGL 120 ICRAQERKKG VPWTEEEHRT FLIGLEKLGK GDWRGISRNY VTTRTPTQVA SHAQKYFLRQ 180 ASLTKKKRRS SLFDMFGSSN NINMETDDPI PTQRDNIIRN NMAIDSSTVP LLEHSIAAKN 240 LNLDHSQKPH SSSTTTRQLP VWIYELTDSQ MKPNSSSSKA SVVPDLELTL AAPTSPNVEQ 300 KNKSSPSPAG PLLNLGTISV T* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 184 | 188 | KKKRR |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Mdo.26737 | 0.0 | leaf | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00565 | DAP | Transfer from AT5G56840 | Download |
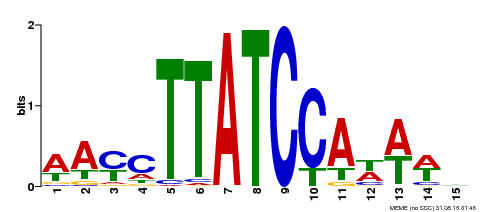
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008387730.1 | 0.0 | transcription factor KUA1-like | ||||
| TrEMBL | A0A498KD06 | 0.0 | A0A498KD06_MALDO; Uncharacterized protein | ||||
| STRING | XP_008387730.1 | 0.0 | (Malus domestica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3213 | 32 | 71 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G56840.1 | 2e-55 | MYB_related family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | MDP0000875621 |



