 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MELO3C002090P1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 292aa MW: 32719.3 Da PI: 6.8541 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 54 | 3.9e-17 | 28 | 75 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg+WT eEd ll++ + ++G g+W++ a++ g++Rt+k+c++rw++yl
MELO3C002090P1 28 RGPWTLEEDNLLIHSISLHGEGRWNLLAKRSGLRRTGKSCRLRWLNYL 75
89********************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 51.1 | 3.2e-16 | 81 | 124 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
rg+ +++E++l++d++ ++G++ W++Ia++++ gRt++++k++w++
MELO3C002090P1 81 RGNLSAQEQLLILDLHSKWGNR-WSKIAQHLP-GRTDNEIKNYWRT 124
7899******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 24.917 | 23 | 79 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.38E-29 | 26 | 122 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.0E-15 | 27 | 77 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 7.6E-16 | 28 | 75 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.0E-21 | 29 | 82 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 6.45E-12 | 30 | 75 | No hit | No description |
| PROSITE profile | PS51294 | 19.748 | 80 | 130 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.8E-14 | 80 | 128 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 9.9E-15 | 81 | 124 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-22 | 83 | 129 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 7.61E-11 | 85 | 124 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 292 aa Download sequence Send to blast |
MSSSSPKSFG SCSEIEDDNS NNGNELRRGP WTLEEDNLLI HSISLHGEGR WNLLAKRSGL 60 RRTGKSCRLR WLNYLKPDVK RGNLSAQEQL LILDLHSKWG NRWSKIAQHL PGRTDNEIKN 120 YWRTRVQKQA RHLNIDTNSE AFQQIIRSYW IPRLIQKINQ SPPSELPITS PPEMASQTSF 180 GFEAPTTAVA QPSAVTPLQI GGDLMGSSCS SSSSSHITQI SSFQNYENFA PFVKDYYHDG 240 GHGGGEMLNW ATTAVAGDSG YPVSHCHVAE NNWMESDFTG YMSNVDELWQ F* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 2e-24 | 25 | 130 | 4 | 108 | B-MYB |
| 1gv2_A | 2e-24 | 25 | 130 | 1 | 105 | MYB PROTO-ONCOGENE PROTEIN |
| 1h8a_C | 5e-24 | 25 | 130 | 24 | 128 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription repressor of phosphate (Pi) starvation-induced genes. Regulates negatively Pi starvation responses via the repression of gibberellic acid (GA) biosynthesis and signaling. Modulates root architecture, phosphatase activity, and Pi uptake and accumulation. {ECO:0000269|PubMed:19529828}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00159 | DAP | Transfer from AT1G25340 | Download |
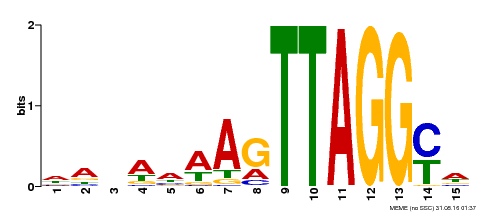
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Slightly induced by salicylic acid (PubMed:16463103). Induced reversibly in response to phosphate (Pi) deficiency but repressed in the presence of Pi, specifically in the leaves. Availability of Pi increases with decreased levels (PubMed:19529828). {ECO:0000269|PubMed:16463103, ECO:0000269|PubMed:19529828}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681932 | 0.0 | LN681932.1 Cucumis melo genomic scaffold, anchoredscaffold00001. | |||
| GenBank | LN713266 | 0.0 | LN713266.1 Cucumis melo genomic chromosome, chr_12. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008458430.1 | 0.0 | PREDICTED: transcription factor MYB108-like | ||||
| Swissprot | Q9C9G7 | 5e-80 | MYB62_ARATH; Transcription factor MYB62 | ||||
| TrEMBL | A0A1S3C8F6 | 0.0 | A0A1S3C8F6_CUCME; transcription factor MYB108-like | ||||
| STRING | XP_008458430.1 | 0.0 | (Cucumis melo) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1639 | 34 | 94 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G25340.1 | 2e-78 | myb domain protein 116 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



