 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MELO3C002649P1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 179aa MW: 20234.1 Da PI: 8.8201 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 121.9 | 2.2e-38 | 56 | 114 | 2 | 60 |
zf-Dof 2 kekalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkk 60
+ek+++cprC+s++tkfCy+nny+++qPr+fCk+C+ryWt+GGalrnvP+G+grrk k
MELO3C002649P1 56 PEKIIPCPRCKSMDTKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPIGAGRRKTKP 114
68999***************************************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD007478 | 2.0E-28 | 55 | 113 | IPR003851 | Zinc finger, Dof-type |
| Pfam | PF02701 | 6.4E-32 | 58 | 113 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 28.36 | 60 | 114 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 62 | 98 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010214 | Biological Process | seed coat development | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:1902455 | Biological Process | negative regulation of stem cell population maintenance | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 179 aa Download sequence Send to blast |
MMGERKHDKN NEDQHGGGIK LFGATIMLQN KRQIIKEEEE EEEANKSDQQ TLEKRPEKII 60 PCPRCKSMDT KFCYFNNYNV NQPRHFCKGC QRYWTAGGAL RNVPIGAGRR KTKPPCRTFG 120 GLPENCVFDS SGIVTVQPFE LEGMVEKWHA VAAAATQGGF RQILPVKRRR DCQDGQTC* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence. Acts as a negative regulator in the phytochrome-mediated light responses. Controls phyB-mediated end-of-day response and the phyA-mediated anthocyanin accumulation. Not involved in direct flowering time regulation. {ECO:0000269|PubMed:19619493}. | |||||
| UniProt | Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence (By similarity). Transcriptional repressor of 'CONSTANS' expression (By similarity). Regulates a photoperiodic flowering response. {ECO:0000250, ECO:0000269|PubMed:19619493}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00166 | DAP | Transfer from AT1G29160 | Download |
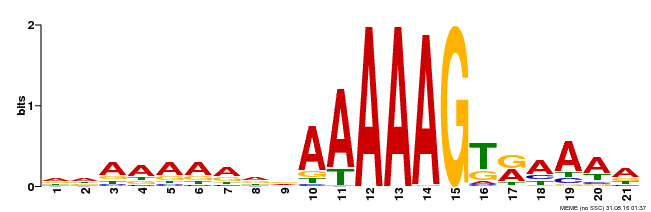
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By red or far-red light. Circadian-regulation. The transcript level rises progressively from dawn and decreases during the night. {ECO:0000269|PubMed:19619493}. | |||||
| UniProt | INDUCTION: Circadian-regulation. The transcript level rises progressively from dawn and decreases during the night. {ECO:0000269|PubMed:19619493}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681932 | 0.0 | LN681932.1 Cucumis melo genomic scaffold, anchoredscaffold00001. | |||
| GenBank | LN713266 | 0.0 | LN713266.1 Cucumis melo genomic chromosome, chr_12. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008443229.1 | 1e-132 | PREDICTED: dof zinc finger protein DOF1.5 | ||||
| Swissprot | O22967 | 5e-57 | CDF4_ARATH; Cyclic dof factor 4 | ||||
| Swissprot | P68350 | 9e-57 | DOF15_ARATH; Dof zinc finger protein DOF1.5 | ||||
| TrEMBL | A0A1S3B895 | 1e-131 | A0A1S3B895_CUCME; dof zinc finger protein DOF1.5 | ||||
| STRING | XP_008443229.1 | 1e-132 | (Cucumis melo) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF6483 | 32 | 51 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G29160.1 | 1e-58 | Dof family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



