 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MELO3C008318P2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 429aa MW: 47357.6 Da PI: 4.5809 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 59 | 1.1e-18 | 84 | 134 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
+ y+GVr++ +g+WvAeIr+p + r r +lg+f ta eAa a+++a++ ++g
MELO3C008318P2 84 CNYRGVRQRT-WGKWVAEIREP---N-RgSRLWLGTFPTAIEAALAYDEAARTMYG 134
56*****999.**********8...3.35***********************9987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 1.13E-30 | 84 | 143 | No hit | No description |
| Pfam | PF00847 | 3.7E-13 | 84 | 134 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.1E-36 | 85 | 148 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 4.71E-20 | 85 | 142 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 21.509 | 85 | 142 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.0E-30 | 85 | 143 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.7E-9 | 86 | 97 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.7E-9 | 108 | 124 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0010286 | Biological Process | heat acclimation | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 429 aa Download sequence Send to blast |
MVEKMGHSDQ YSDAISLPTN SIRKRKSRSR RDRSTVAETL AKWKAYNECF DSSNDGGKLI 60 RKAPAKGSKK GCMKGKGGPL NSHCNYRGVR QRTWGKWVAE IREPNRGSRL WLGTFPTAIE 120 AALAYDEAAR TMYGQTARLN LPNIKNRGQL QGILLEDYLG LRNSDSSTTT STCSESTTTT 180 SNQSEVCVPE EFTVRPQLVP LNVKSEDGEG ESRTGDHGDE TATPMCLENQ VKHEDCNGQT 240 AALVAEFPCL DQLQNFQMAE IFEPRTGPQT GPMLTPLSLE KQVKDEDLDA VYRERSDDQA 300 VLSEAGISSL YDLQNFEMDE MFDVEELLSL ISSDSLHDPT NILKGNADGY TSMVPSQVGS 360 IGSEKPPNSS YQMQNPDAKL LGSPQQMERT PADVDYGFDF LKQGREEDLN AAADDCVRYL 420 NEIGDLGF* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 9e-18 | 85 | 142 | 2 | 60 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| 2gcc_A | 9e-18 | 85 | 149 | 5 | 70 | ATERF1 |
| 3gcc_A | 9e-18 | 85 | 149 | 5 | 70 | ATERF1 |
| 5wx9_A | 5e-17 | 74 | 148 | 3 | 78 | Ethylene-responsive transcription factor ERF096 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 24 | 31 | RKSRSRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00302 | DAP | Transfer from AT2G40340 | Download |
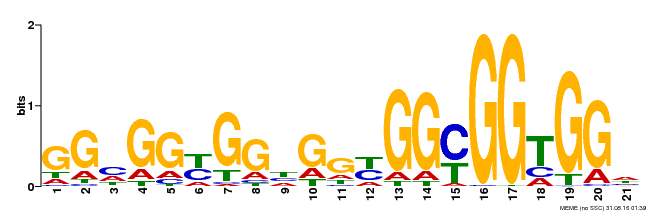
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681815 | 0.0 | LN681815.1 Cucumis melo genomic scaffold, anchoredscaffold00008. | |||
| GenBank | LN713257 | 0.0 | LN713257.1 Cucumis melo genomic chromosome, chr_3. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008441482.1 | 0.0 | PREDICTED: dehydration-responsive element-binding protein 2C isoform X1 | ||||
| TrEMBL | A0A1S3B4B1 | 0.0 | A0A1S3B4B1_CUCME; dehydration-responsive element-binding protein 2C isoform X1 | ||||
| STRING | XP_008441482.1 | 0.0 | (Cucumis melo) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1175 | 34 | 106 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G40340.1 | 4e-32 | ERF family protein | ||||



