 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MELO3C010949P1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | ZF-HD | ||||||||
| Protein Properties | Length: 315aa MW: 33766.4 Da PI: 7.0188 | ||||||||
| Description | ZF-HD family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | ZF-HD_dimer | 102.8 | 2.2e-32 | 84 | 140 | 3 | 60 |
ZF-HD_dimer 3 kvrYkeClkNhAaslGghavDGCgEfmpsegeegtaaalkCaACgCHRnFHRreveee 60
+vrY+eClkNhAas+Gg+ +DGCgEfmps ge+gt +alkCaAC+CHRnFHR+e + e
MELO3C010949P1 84 GVRYRECLKNHAASVGGNIYDGCGEFMPS-GEDGTLEALKCAACECHRNFHRKEIDGE 140
79**************************9.999********************98865 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04770 | 7.8E-30 | 85 | 137 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| ProDom | PD125774 | 2.0E-23 | 85 | 138 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| TIGRFAMs | TIGR01566 | 1.7E-30 | 86 | 137 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| PROSITE profile | PS51523 | 26.487 | 87 | 136 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| Gene3D | G3DSA:1.10.10.60 | 2.5E-31 | 220 | 292 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 2.5E-20 | 222 | 292 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01565 | 2.1E-28 | 230 | 287 | IPR006455 | Homeodomain, ZF-HD class |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 315 aa Download sequence Send to blast |
MDMREQEKEV IMPGANSGAG AYGHIFGETS SDRIHNGSHH HIQTDHHDAG ENVGGGGGGG 60 GGGGGGGGNF SSGGRSKARG GVSGVRYREC LKNHAASVGG NIYDGCGEFM PSGEDGTLEA 120 LKCAACECHR NFHRKEIDGE TQLNISPNYR RGLMLNHLQL PPPLPSPSAL HGHHKFSMAL 180 NLHSSPTAPI IAPMNVAFAG GGGNESSSED LNVFHSNAEV MPPSSFSLSK KRFRTKFTQE 240 QKDRMLEFAE KVGWRIQKQD EEEVERFCTE VGVKRQVLKV WMHNNKNTVK KQNENHEPDQ 300 LTGTGAGAGI TNES* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wh7_A | 4e-25 | 229 | 288 | 16 | 75 | ZF-HD homeobox family protein |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
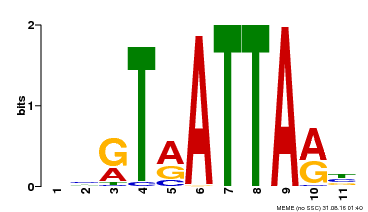
| UniProt | Putative transcription factor. Binds DNA at 5'-ATTA-3' consensus promoter regions. Regulates floral architecture and leaf development. Regulators in the abscisic acid (ABA) signal pathway that confers sensitivity to ABA in an ARF2-dependent manner. {ECO:0000269|PubMed:16428600, ECO:0000269|PubMed:21059647, ECO:0000269|PubMed:21779177}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00238 | DAP | Transfer from AT1G75240 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by abscisic acid (ABA) and ARF2. {ECO:0000269|PubMed:21779177}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681823 | 0.0 | LN681823.1 Cucumis melo genomic scaffold, anchoredscaffold00014. | |||
| GenBank | LN713257 | 0.0 | LN713257.1 Cucumis melo genomic chromosome, chr_3. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008444722.1 | 0.0 | PREDICTED: zinc-finger homeodomain protein 2 | ||||
| Swissprot | Q9FRL5 | 2e-66 | ZHD5_ARATH; Zinc-finger homeodomain protein 5 | ||||
| TrEMBL | A0A1S3BAZ3 | 0.0 | A0A1S3BAZ3_CUCME; zinc-finger homeodomain protein 2 | ||||
| STRING | XP_008444722.1 | 0.0 | (Cucumis melo) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF206 | 34 | 253 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G18350.1 | 1e-62 | homeobox protein 24 | ||||



