 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MELO3C013970P1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | YABBY | ||||||||
| Protein Properties | Length: 185aa MW: 20476.3 Da PI: 8.6198 | ||||||||
| Description | YABBY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | YABBY | 213 | 9.6e-66 | 15 | 181 | 5 | 169 |
YABBY 5 ssseqvCyvqCnfCntilavsvPstslfkvvtvrCGhCtsllsvnlakasqllaaeshldeslkee..lleelkveeenlksnvekeesastsvs 97
s+q+Cyv+Cn C+t+lavsvPstslfk vtvrCG+C++ll v++ ll++ s+++ ++ + ++++ e+++ + + ++t+
MELO3C013970P1 15 PPSDQLCYVHCNICDTVLAVSVPSTSLFKRVTVRCGYCANLLPVSMCGG-MLLPSPSQFHGFTHSTtfFSPNTHNFLEEISNPNPNFLMNQTEGI 108
679****************************************987665.566777764433333323222222222222222222223333334 PP
YABBY 98 seklsenedeevprvpp.virPPekrqrvPsaynrfikeeiqrikasnPdishreafsaaaknWahfPkihfg 169
++ + ++vpr+pp ++rPPekrqrvPsaynrfik+eiqrika+nPdishreafsaaaknWahfP+i fg
MELO3C013970P1 109 DLTMATRVPNDVPRQPPtINRPPEKRQRVPSAYNRFIKDEIQRIKAANPDISHREAFSAAAKNWAHFPHIRFG 181
44568888999**999989****************************************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04690 | 2.4E-65 | 16 | 181 | IPR006780 | YABBY protein |
| SuperFamily | SSF47095 | 8.77E-9 | 120 | 175 | IPR009071 | High mobility group box domain |
| Gene3D | G3DSA:1.10.30.10 | 8.6E-5 | 130 | 176 | IPR009071 | High mobility group box domain |
| CDD | cd01390 | 1.30E-5 | 136 | 172 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009909 | Biological Process | regulation of flower development | ||||
| GO:0009933 | Biological Process | meristem structural organization | ||||
| GO:0009944 | Biological Process | polarity specification of adaxial/abaxial axis | ||||
| GO:0010093 | Biological Process | specification of floral organ identity | ||||
| GO:0010154 | Biological Process | fruit development | ||||
| GO:0010158 | Biological Process | abaxial cell fate specification | ||||
| GO:0010159 | Biological Process | specification of organ position | ||||
| GO:0010450 | Biological Process | inflorescence meristem growth | ||||
| GO:1902183 | Biological Process | regulation of shoot apical meristem development | ||||
| GO:2000024 | Biological Process | regulation of leaf development | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 185 aa Download sequence Send to blast |
MSSSSLPTTF SQLPPPSDQL CYVHCNICDT VLAVSVPSTS LFKRVTVRCG YCANLLPVSM 60 CGGMLLPSPS QFHGFTHSTT FFSPNTHNFL EEISNPNPNF LMNQTEGIDL TMATRVPNDV 120 PRQPPTINRP PEKRQRVPSA YNRFIKDEIQ RIKAANPDIS HREAFSAAAK NWAHFPHIRF 180 GSSS* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Seems to be associated with phloem cell differentiation. {ECO:0000269|PubMed:17676337}. | |||||
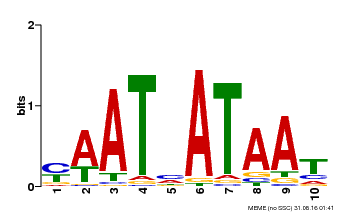
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00620 | PBM | Transfer from AT2G45190 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681860 | 3e-91 | LN681860.1 Cucumis melo genomic scaffold, anchoredscaffold00021. | |||
| GenBank | LN713260 | 3e-91 | LN713260.1 Cucumis melo genomic chromosome, chr_6. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008448889.1 | 1e-135 | PREDICTED: protein YABBY 4-like | ||||
| Refseq | XP_016900570.1 | 1e-135 | PREDICTED: protein YABBY 4-like | ||||
| Swissprot | A2X7Q3 | 1e-59 | YAB4_ORYSI; Protein YABBY 4 | ||||
| Swissprot | Q6H668 | 1e-59 | YAB4_ORYSJ; Protein YABBY 4 | ||||
| TrEMBL | A0A1S4DXY3 | 1e-134 | A0A1S4DXY3_CUCME; protein YABBY 4-like | ||||
| STRING | XP_008448889.1 | 1e-134 | (Cucumis melo) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2040 | 34 | 90 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G45190.1 | 7e-57 | YABBY family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



