 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MELO3C016213P1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 669aa MW: 75113.5 Da PI: 6.2484 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 155.9 | 2.9e-48 | 59 | 194 | 1 | 134 |
DUF822 1 ggsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspesslq 95
gg+ +++++kE+E++k+RER+RRai+++i+aGLR++Gn++lp+raD+n+Vl+AL+reAGwvve+DGttyr+++ p ++++ ++ +s es+++
MELO3C016213P1 59 GGKAKREREKEKERTKLRERHRRAITSRILAGLRQYGNFPLPARADMNDVLAALAREAGWVVEADGTTYRQSTPPS--QSQGAAFPVRSGESPIS 151
57899*****************************************************************999888..66677788888888888 PP
DUF822 96 .sslkssalaspvesysaspksssfpspssldsislasa...a 134
s k +++++ ++ + ++++ ++++sp+slds+ +++ +
MELO3C016213P1 152 sGSFKGCSIKAALDCQPSVLRIDESLSPASLDSVVITERdakN 194
7789******************************999875442 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 8.0E-49 | 60 | 204 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| SuperFamily | SSF51445 | 8.44E-160 | 226 | 660 | IPR017853 | Glycoside hydrolase superfamily |
| Gene3D | G3DSA:3.20.20.80 | 6.2E-171 | 229 | 660 | IPR013781 | Glycoside hydrolase, catalytic domain |
| Pfam | PF01373 | 3.9E-81 | 253 | 624 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.3E-53 | 266 | 280 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.3E-53 | 287 | 305 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.3E-53 | 309 | 330 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.3E-53 | 402 | 424 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.3E-53 | 475 | 494 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.3E-53 | 509 | 525 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.3E-53 | 526 | 537 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.3E-53 | 544 | 567 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.3E-53 | 582 | 604 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 669 aa Download sequence Send to blast |
MSGSLNDDSF HQDLQSQANH ASDYLSHQLQ PPPPPRRPRG FAATAAAMGP TTTTAATTGG 60 KAKREREKEK ERTKLRERHR RAITSRILAG LRQYGNFPLP ARADMNDVLA ALAREAGWVV 120 EADGTTYRQS TPPSQSQGAA FPVRSGESPI SSGSFKGCSI KAALDCQPSV LRIDESLSPA 180 SLDSVVITER DAKNEKYTAL SPLNAAHCLE DQLIQDIRCR ENESQFRGTP YVPVYVMLAT 240 GFINNFCQLI DPDGVRQELS HLQSLNVDGV IVDCWWGIVE AWNPQKYVWS GYRDLFNIIR 300 EFKLKVQVVM AFHASGGTES GDAFIKLPQW VLEIGKENPD IFFTDREGRR NKDCLSWGID 360 KERVLRGRTG IEVYFDFMRS FHTEFNDLFA EGLVSAIEVG LGASGELKYP SFSERMGWRY 420 PGIGEFQCYD KYLQQSLRKA AGLRGHSFWA RGPDNAGQYN SRPHETGFFC ERGDYDSYYG 480 RFFLQWYAQT LIYHVDNVLS LASLVFEETK FIVKIPAVYW WYKTSSHAAE LTSGFYNPSN 540 QDGYSPVFDV LKKHSVIVKL VCCGMPVAGQ DVDDALADPE SLSWQILNSA WDRGLTVAGE 600 NSLSCYDRDG YVRIIDMAKP RSDPDRRHFS FFAYRQPSAL IQGAVCFPEL DYFIKCMHGE 660 IEGDIVPS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wdq_A | 1e-123 | 231 | 660 | 11 | 444 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
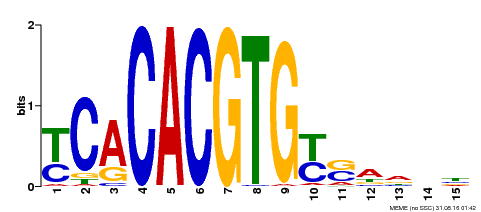
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681873 | 0.0 | LN681873.1 Cucumis melo genomic scaffold, anchoredscaffold00027. | |||
| GenBank | LN713261 | 0.0 | LN713261.1 Cucumis melo genomic chromosome, chr_7. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008451866.1 | 0.0 | PREDICTED: beta-amylase 8 | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | A0A1S3BTA5 | 0.0 | A0A1S3BTA5_CUCME; Beta-amylase | ||||
| STRING | XP_008451866.1 | 0.0 | (Cucumis melo) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF11902 | 30 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.2 | 0.0 | beta-amylase 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



