 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MELO3C019923P1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 554aa MW: 61221.7 Da PI: 5.6181 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 59.4 | 8e-19 | 38 | 85 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT Ede+l+++vk++G g+W+++ ++ g+ R++k+c++rw ++l
MELO3C019923P1 38 KGPWTSAEDEILIEYVKKHGEGNWNAVQKHSGLSRCGKSCRLRWANHL 85
79******************************************9986 PP
| |||||||
| 2 | Myb_DNA-binding | 52.1 | 1.5e-16 | 91 | 134 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
+g++T+eE+ l++++++++G++ W++ a++++ gRt++++k++w++
MELO3C019923P1 91 KGAFTAEEEHLIIELHAKMGNK-WARMAAHLP-GRTDNEIKNYWNT 134
799*******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 17.984 | 33 | 85 | IPR017930 | Myb domain |
| SMART | SM00717 | 9.2E-17 | 37 | 87 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 3.67E-31 | 37 | 132 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 1.0E-16 | 38 | 85 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 9.7E-25 | 39 | 93 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 8.84E-13 | 40 | 85 | No hit | No description |
| PROSITE profile | PS51294 | 26.06 | 86 | 140 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.1E-15 | 90 | 138 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.4E-15 | 91 | 134 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.64E-11 | 93 | 134 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 2.6E-25 | 94 | 139 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009789 | Biological Process | positive regulation of abscisic acid-activated signaling pathway | ||||
| GO:0043068 | Biological Process | positive regulation of programmed cell death | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0045926 | Biological Process | negative regulation of growth | ||||
| GO:0048235 | Biological Process | pollen sperm cell differentiation | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 554 aa Download sequence Send to blast |
MRHPKNEIED NLPSQDQTLS PLLDEDSGGN ASGIVLKKGP WTSAEDEILI EYVKKHGEGN 60 WNAVQKHSGL SRCGKSCRLR WANHLRPNLK KGAFTAEEEH LIIELHAKMG NKWARMAAHL 120 PGRTDNEIKN YWNTRIKRRQ RAGLPLYPPE VCLRTWQALQ QTQDSGGSTI VDNDHHDLLR 180 ANSYDIPDVT FHSLKPQSAL SYMPELPDIS SCMLKRGLDS SQYCNFMQSA LHRQKRFRDS 240 ASLFPGPDGS VKTPFHQFED NSYSQAAQSF GTPFAVESNP TTKNAMSFGS FEGSHSLTNG 300 NSSASQHSKE TEKLELPSLQ YPETDLTSWD TTIQPAMFES VDPFIQSTPT FVLQATDRQS 360 PCHSGLLEAL VYPKPMGPKN HPSDKNSNSC SVTPGDVTDS YNMAASKTEI DDYTEVISPF 420 GHSTSSLFSE CTPISATGSS YEDPTLTEAF SGSHVKSEPL DHAWTPDREK AAKSRVNFAR 480 PDALLTSDWH DRSSGIVEDT TNVTDAISLL LGDDLAADYE HFPNGISTTH SAWGLDSCSW 540 NNMPAVCHMS DLP* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 5e-32 | 36 | 139 | 5 | 107 | B-MYB |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00490 | DAP | Transfer from AT5G06100 | Download |
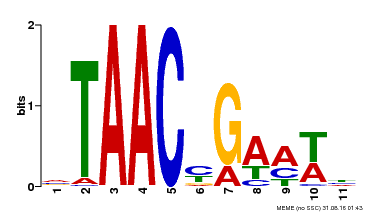
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681821 | 0.0 | LN681821.1 Cucumis melo genomic scaffold, anchoredscaffold00040. | |||
| GenBank | LN713257 | 0.0 | LN713257.1 Cucumis melo genomic chromosome, chr_3. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008456638.1 | 0.0 | PREDICTED: transcription factor GAMYB | ||||
| Refseq | XP_008456639.1 | 0.0 | PREDICTED: transcription factor GAMYB | ||||
| TrEMBL | A0A1S3C3Q3 | 0.0 | A0A1S3C3Q3_CUCME; transcription factor GAMYB | ||||
| STRING | XP_008456638.1 | 0.0 | (Cucumis melo) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2047 | 34 | 85 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G06100.3 | 1e-120 | myb domain protein 33 | ||||



