 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MELO3C019954P2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 453aa MW: 51280.7 Da PI: 4.3825 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 160.2 | 8e-50 | 17 | 143 | 2 | 128 |
NAM 2 ppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLp.k.kvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevls 94
ppGfrFhPtdeel+ +yLk k+ ++kl+l ++i+e+d+yk+eP +Lp + k+k+++++w+fFs+r+++y++ +r +rat+sgyWkatgkd+ + +
MELO3C019954P2 17 PPGFRFHPTDEELILYYLKFKICRRKLKL-DIIRETDVYKWEPDELPgQsKLKTGDRQWFFFSPRERRYPNASRLSRATRSGYWKATGKDRIIQC 110
8****************************.99**************963589999**************************************99 PP
NAM 95 kkgelvglkktLvfykgrapkgektdWvmheyrl 128
++++vg+kktLvfy grap+ge+tdWvmhey+l
MELO3C019954P2 111 -NSRNVGVKKTLVFYLGRAPNGERTDWVMHEYTL 143
.999****************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 4.71E-57 | 15 | 166 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 53.355 | 16 | 166 | IPR003441 | NAC domain |
| Pfam | PF02365 | 3.6E-26 | 17 | 143 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0070301 | Biological Process | cellular response to hydrogen peroxide | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005789 | Cellular Component | endoplasmic reticulum membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 453 aa Download sequence Send to blast |
MSTASSSKGL CDEGDWPPGF RFHPTDEELI LYYLKFKICR RKLKLDIIRE TDVYKWEPDE 60 LPGQSKLKTG DRQWFFFSPR ERRYPNASRL SRATRSGYWK ATGKDRIIQC NSRNVGVKKT 120 LVFYLGRAPN GERTDWVMHE YTLDEDELKR CKNVKDYYAL YKLYKKSGPG PKNGEQYGAP 180 FREEDWVDDA DCLEPQVKTV DEVDSVVRER DNGQIQLSSE DIEELMKQIV NDPVLELPSV 240 SGYHQLGSAL QVDDKEETAS TMIDAYAQEH ILPQADKVYH FSVQPSDLHA SFDFTQSGIS 300 QLQSFEAEVS SALKDCEEEG DFLEIYDLIG PEPTPVANVN PLGNIPPSEL DGLSELDLFH 360 DANMFLRDLG PITTETVLDP YLNALDVDVA NNLNGNLQHV SYQQNQTDDG FWKNNETENA 420 FSFHESHGQF VTQSTLVAGS ISSSTQSSCS DE* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 1e-45 | 17 | 166 | 18 | 165 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 1e-45 | 17 | 166 | 18 | 165 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 1e-45 | 17 | 166 | 18 | 165 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 1e-45 | 17 | 166 | 18 | 165 | NO APICAL MERISTEM PROTEIN |
| 4dul_A | 1e-45 | 17 | 166 | 18 | 165 | NAC domain-containing protein 19 |
| 4dul_B | 1e-45 | 17 | 166 | 18 | 165 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator activated by proteolytic cleavage through regulated intramembrane proteolysis (RIP). Transcriptional activator that acts as positive regulator of AOX1A during mitochondrial dysfunction. Binds directly to AOX1A promoter. Mediates mitochondrial retrograde signaling. {ECO:0000269|PubMed:24045017}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00179 | DAP | Transfer from AT1G34190 | Download |
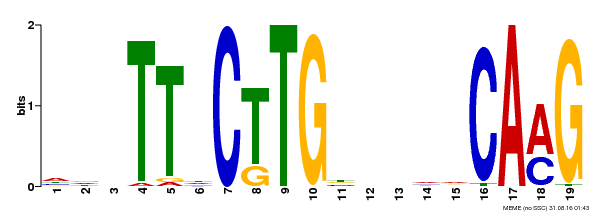
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By cold and drought stresses. {ECO:0000269|PubMed:17158162}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681821 | 0.0 | LN681821.1 Cucumis melo genomic scaffold, anchoredscaffold00040. | |||
| GenBank | LN713257 | 0.0 | LN713257.1 Cucumis melo genomic chromosome, chr_3. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008456695.1 | 0.0 | PREDICTED: NAC domain-containing protein 17-like | ||||
| Swissprot | Q9XIC5 | 1e-107 | NAC17_ARATH; NAC domain-containing protein 17 | ||||
| TrEMBL | A0A1S3C3X9 | 0.0 | A0A1S3C3X9_CUCME; NAC domain-containing protein 17-like | ||||
| STRING | XP_008456695.1 | 0.0 | (Cucumis melo) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G34190.1 | 2e-98 | NAC domain containing protein 17 | ||||



