 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MELO3C021144P1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 298aa MW: 32888.4 Da PI: 8.2487 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 130 | 8.7e-41 | 161 | 238 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cq+egC+adls+ak+yhrrhkvCe+hska++v+++gl+qrfCqqCsrfh + efD++krsCr+rLa+hn+rrrk+q+
MELO3C021144P1 161 RCQAEGCNADLSQAKHYHRRHKVCEFHSKASTVITAGLTQRFCQQCSRFHVVGEFDNGKRSCRKRLADHNRRRRKTQQ 238
6**************************************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 2.8E-34 | 156 | 223 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 31.573 | 159 | 236 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.83E-37 | 160 | 240 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 1.2E-30 | 162 | 235 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009554 | Biological Process | megasporogenesis | ||||
| GO:0009556 | Biological Process | microsporogenesis | ||||
| GO:0048653 | Biological Process | anther development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 298 aa Download sequence Send to blast |
MLDYEWGSTN PPPIILSGEE SEPVFNSDPS HSHNFPTFID LHHHNTFFTP PPPPPPPPPS 60 TAAFSSHLGG FYDHPQSYTM GTSSYHHYNS VTGGGGEFML VPKSEDVCGS MAAPVEFAAR 120 IGLNLGGRTY FSSEEEEFVN RLYRKTRSGS EMGGSGNWAV RCQAEGCNAD LSQAKHYHRR 180 HKVCEFHSKA STVITAGLTQ RFCQQCSRFH VVGEFDNGKR SCRKRLADHN RRRRKTQQSH 240 HQGTPNSSQL LQTTPSSSTH SAATRSTMES TGQSAWSVTV AVSPTRMSLD CLNQRPF* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 4e-32 | 153 | 235 | 1 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
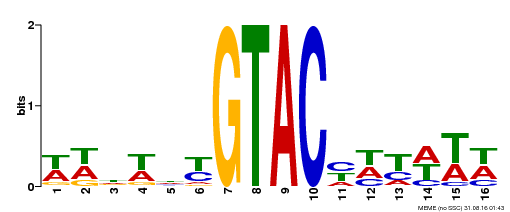
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. Binds specifically to the 5'-GTAC-3' core sequence. Involved in development and floral organogenesis. Required for ovule differentiation, pollen production, filament elongation, seed formation and siliques elongation. Also seems to play a role in the formation of trichomes on sepals. May positively modulate gibberellin (GA) signaling in flower. {ECO:0000269|PubMed:12671094, ECO:0000269|PubMed:16095614, ECO:0000269|PubMed:17093870}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00090 | SELEX | Transfer from AT1G02065 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681924 | 0.0 | LN681924.1 Cucumis melo genomic scaffold, anchoredscaffold00047. | |||
| GenBank | LN713265 | 0.0 | LN713265.1 Cucumis melo genomic chromosome, chr_11. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008458127.1 | 0.0 | PREDICTED: squamosa promoter-binding-like protein 8 | ||||
| Refseq | XP_008458128.1 | 0.0 | PREDICTED: squamosa promoter-binding-like protein 8 | ||||
| Swissprot | Q8GXL3 | 5e-63 | SPL8_ARATH; Squamosa promoter-binding-like protein 8 | ||||
| TrEMBL | A0A1S3C8D3 | 0.0 | A0A1S3C8D3_CUCME; squamosa promoter-binding-like protein 8 | ||||
| STRING | XP_008458127.1 | 0.0 | (Cucumis melo) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF6801 | 33 | 48 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G02065.1 | 5e-67 | squamosa promoter binding protein-like 8 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



