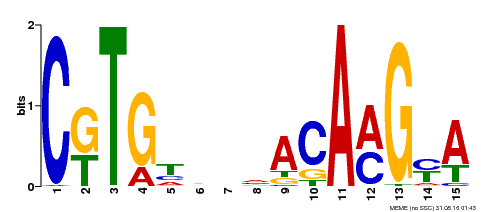
| Signature Domain? help Back to Top |
 |
| No. |
Domain |
Score |
E-value |
Start |
End |
HMM Start |
HMM End |
| 1 | NAM | 176.8 | 5.9e-55 | 24 | 149 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevlsk 95
lppGfrFhPtdeel+++yL +kv + +++ +i ++d++k+ePwdLp ++k +ekewyfF+ +d+ky+tg r+nrat+sgyWkatgkdke+++
MELO3C022002P1 24 LPPGFRFHPTDEELITHYLSPKVLNPNFSS-IAIGHADLNKSEPWDLPGRAKMGEKEWYFFCVKDRKYPTGLRTNRATDSGYWKATGKDKEIFR- 116
79*************************999.78***************99999*****************************************. PP
NAM 96 kgelvglkktLvfykgrapkgektdWvmheyrl 128
+++lvg+kktLvfykgrapkgek++Wvmhe+rl
MELO3C022002P1 117 GKKLVGMKKTLVFYKGRAPKGEKSNWVMHEFRL 149
99*****************************98 PP
|
| Gene Ontology ? help Back to Top |
| GO Term |
GO Category |
GO Description |
| GO:0009651 | Biological Process | response to salt stress |
| GO:0009723 | Biological Process | response to ethylene |
| GO:0009733 | Biological Process | response to auxin |
| GO:0009737 | Biological Process | response to abscisic acid |
| GO:0010029 | Biological Process | regulation of seed germination |
| GO:0042542 | Biological Process | response to hydrogen peroxide |
| GO:0043068 | Biological Process | positive regulation of programmed cell death |
| GO:0048527 | Biological Process | lateral root development |
| GO:0051091 | Biological Process | positive regulation of sequence-specific DNA binding transcription factor activity |
| GO:0090400 | Biological Process | stress-induced premature senescence |
| GO:1900057 | Biological Process | positive regulation of leaf senescence |
| GO:1902074 | Biological Process | response to salt |
| GO:1904250 | Biological Process | positive regulation of age-related resistance |
| GO:0005634 | Cellular Component | nucleus |
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding |
| GO:0042803 | Molecular Function | protein homodimerization activity |
| GO:0043565 | Molecular Function | sequence-specific DNA binding |
| Functional Description ? help
Back to Top |
| Source |
Description |
| UniProt | Transcription activator that binds to DNA in promoters of target genes on a specific bipartite motif 5'-[ACG][CA]GT[AG](5-6n)[CT]AC[AG]-3' (PubMed:23340744). Promotes lateral root development (PubMed:16359384). Triggers the expression of senescence-associated genes during age-, salt- and dark-induced senescence through a regulatory network that may involve cross-talk with salt- and H(2)O(2)-dependent signaling pathways (PubMed:9351240, PubMed:15295076, PubMed:20113437, PubMed:21303842). Regulates also genes during seed germination (PubMed:20113437). Regulates positively aging-induced cell death (PubMed:19229035). Involved in age-related resistance (ARR) against Pseudomonas syringae pv. tomato and Hyaloperonospora arabidopsidis (PubMed:19694953). Antagonizes GLK1 and GLK2 transcriptional activity, shifting the balance from chloroplast maintenance towards deterioration during leaf senescence (PubMed:23459204). Promotes the expression of senescence-associated genes, including ENDO1/BFN1, SWEET15/SAG29 and SINA1/At3g13672, during senescence onset (PubMed:23340744). {ECO:0000269|PubMed:15295076, ECO:0000269|PubMed:16359384, ECO:0000269|PubMed:19229035, ECO:0000269|PubMed:19694953, ECO:0000269|PubMed:20113437, ECO:0000269|PubMed:21303842, ECO:0000269|PubMed:23340744, ECO:0000269|PubMed:23459204, ECO:0000269|PubMed:9351240}. |
| Regulation -- Description ? help
Back to Top |
| Source |
Description |
| UniProt | INDUCTION: High levels during senescence (e.g. age-, salt- and dark-related) (PubMed:19229035, PubMed:20113437, PubMed:21511905, PubMed:22930749). By salt stress in an ethylene- and auxin-dependent manner (PubMed:16359384, PubMed:19608714, PubMed:20113437, PubMed:20404534). Induced by H(2)O(2) (PubMed:20404534). Accumulates in response to abscisic acid (ABA), ethylene (ACC) and auxin (NAA) (PubMed:16359384, PubMed:19608714). Repressed by high auxin (IAA) levels (PubMed:21511905). Age-related resistance (ARR)-associated accumulation (PubMed:19694953). Repressed by miR164 (PubMed:19229035). {ECO:0000269|PubMed:16359384, ECO:0000269|PubMed:19229035, ECO:0000269|PubMed:19608714, ECO:0000269|PubMed:19694953, ECO:0000269|PubMed:20113437, ECO:0000269|PubMed:20404534, ECO:0000269|PubMed:21511905, ECO:0000269|PubMed:22930749}. |