 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MELO3C022962P1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 320aa MW: 36685.4 Da PI: 8.4483 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 161.3 | 3.7e-50 | 21 | 157 | 2 | 128 |
NAM 2 ppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevlsk. 95
pGfrFhPtdeelv +yL++kve+k++ + e+ik++diyk++PwdLpk + ++kewyfF+ r +ky+++ r+nr+t sg+Wkatg dk+++s+
MELO3C022962P1 21 LPGFRFHPTDEELVGFYLRRKVEKKPIAI-EIIKQIDIYKYDPWDLPKVSNVGDKEWYFFCIRGRKYRNSIRPNRVTGSGFWKATGIDKPIFSTl 114
69***************************.89***************888889****************************************99 PP
NAM 96 ..........kgelvglkktLvfykgrapkgektdWvmheyrl 128
+++++glkk+Lv+y+g+a kg+ktdW+mhe+rl
MELO3C022962P1 115 klssndnannNNNVIGLKKSLVYYRGSAGKGTKTDWMMHEFRL 157
99988887776777***************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 4.32E-55 | 19 | 184 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 53.733 | 20 | 185 | IPR003441 | NAC domain |
| Pfam | PF02365 | 5.5E-25 | 22 | 157 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005992 | Biological Process | trehalose biosynthetic process | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0006561 | Biological Process | proline biosynthetic process | ||||
| GO:0009718 | Biological Process | anthocyanin-containing compound biosynthetic process | ||||
| GO:0010120 | Biological Process | camalexin biosynthetic process | ||||
| GO:0042538 | Biological Process | hyperosmotic salinity response | ||||
| GO:1900056 | Biological Process | negative regulation of leaf senescence | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 320 aa Download sequence Send to blast |
MEVTSKLMDI VVDHEDDQII LPGFRFHPTD EELVGFYLRR KVEKKPIAIE IIKQIDIYKY 60 DPWDLPKVSN VGDKEWYFFC IRGRKYRNSI RPNRVTGSGF WKATGIDKPI FSTLKLSSND 120 NANNNNNVIG LKKSLVYYRG SAGKGTKTDW MMHEFRLPFN NNNIIPHDQD HNNAEVWTLC 180 RIFKRIPSFK KYIPTHNNNN NNNQTYPKPQ NPNNSNIIFT SKPKTNFQSQ PYSTDIHHLI 240 HLHETKPKPF EQLTNPNLTL PSCANSSFES SPICLSSLWN GVEGGGGGDD NFTTSGGWDE 300 LRPIVQFAVD PSFQLYECI* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3ulx_A | 3e-47 | 13 | 191 | 8 | 174 | Stress-induced transcription factor NAC1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds to the 5'- RRYGCCGT-3' consensus core sequence. Central longevity regulator. Negative regulator of leaf senescence. Modulates cellular H(2)O(2) levels and enhances tolerance to various abiotic stresses through the regulation of DREB2A. {ECO:0000269|PubMed:22345491}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00310 | DAP | Transfer from AT2G43000 | Download |
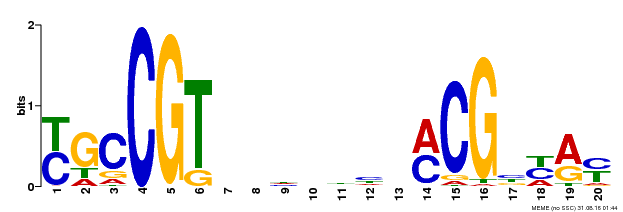
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by H(2)O(2), paraquat, ozone, 3-aminotriazole and salt stress. {ECO:0000269|PubMed:22345491}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681865 | 0.0 | LN681865.1 Cucumis melo genomic scaffold, anchoredscaffold01599. | |||
| GenBank | LN713261 | 0.0 | LN713261.1 Cucumis melo genomic chromosome, chr_7. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008466794.1 | 0.0 | PREDICTED: transcription factor JUNGBRUNNEN 1-like isoform X1 | ||||
| Swissprot | Q9SK55 | 3e-87 | NAC42_ARATH; Transcription factor JUNGBRUNNEN 1 | ||||
| TrEMBL | A0A1S3CTC4 | 0.0 | A0A1S3CTC4_CUCME; transcription factor JUNGBRUNNEN 1-like isoform X1 | ||||
| STRING | XP_008466794.1 | 0.0 | (Cucumis melo) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1013 | 34 | 112 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G43000.1 | 2e-86 | NAC domain containing protein 42 | ||||



