 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MELO3C023549P1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 294aa MW: 33657.4 Da PI: 5.6746 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 47 | 6e-15 | 18 | 65 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g W++eEde+l ++ G g+W+ Iar g+ R++k+c++rw +yl
MELO3C023549P1 18 KGLWSPEEDEKLMRYMLSNGQGCWTDIARNAGLLRCGKSCRLRWINYL 65
678*****************************99************97 PP
| |||||||
| 2 | Myb_DNA-binding | 49.8 | 7.7e-16 | 71 | 115 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
rg+++++E++l+++ + +lG++ W+ Ia++++ gRt++++k++w++
MELO3C023549P1 71 RGAFSPQEEQLILHFHSLLGNR-WSQIAARLP-GRTDNEIKNFWNST 115
89********************.*********.***********986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 16.738 | 13 | 65 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 4.12E-28 | 15 | 112 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.7E-11 | 17 | 67 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.1E-13 | 18 | 65 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.2E-22 | 19 | 72 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 4.95E-9 | 21 | 65 | No hit | No description |
| PROSITE profile | PS51294 | 24.973 | 66 | 120 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.4E-15 | 70 | 118 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 6.3E-15 | 71 | 115 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.0E-25 | 73 | 121 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.54E-10 | 73 | 116 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:2000652 | Biological Process | regulation of secondary cell wall biogenesis | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 294 aa Download sequence Send to blast |
MRKPAECSTA AANIRLRKGL WSPEEDEKLM RYMLSNGQGC WTDIARNAGL LRCGKSCRLR 60 WINYLRPDLK RGAFSPQEEQ LILHFHSLLG NRWSQIAARL PGRTDNEIKN FWNSTLKKRM 120 KNNRNYNIIP SPNLSHSSEL PRSSSNSFGE QSSSSKPLLF STFSDDNYQL FNTFGGGGSG 180 GGDGITEEGY FGDYTTTTIV EPKDFVRDFY DLSHLHQLEN KDIIIEEINK VPSMNNNPQQ 240 NNYELFNNID QMSFEVEDIF GNNNNCQNNQ LQDHFKIGAN WDFDPFILDF QID* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 9e-28 | 16 | 120 | 5 | 108 | B-MYB |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00336 | DAP | Transfer from AT3G08500 | Download |
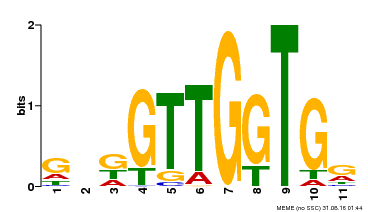
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681804 | 0.0 | LN681804.1 Cucumis melo genomic scaffold, anchoredscaffold00060. | |||
| GenBank | LN713255 | 0.0 | LN713255.1 Cucumis melo genomic chromosome, chr_1. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008461287.1 | 0.0 | PREDICTED: transcription factor MYB46 | ||||
| TrEMBL | A0A1S3CED2 | 0.0 | A0A1S3CED2_CUCME; transcription factor MYB46 | ||||
| STRING | XP_008461287.1 | 0.0 | (Cucumis melo) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF774 | 34 | 128 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G12870.1 | 4e-69 | myb domain protein 46 | ||||



