 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MELO3C024799P1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 285aa MW: 30740.4 Da PI: 8.4722 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 60.5 | 3.7e-19 | 9 | 54 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+g+W++eEd+ l ++v+++G+++W++I++ ++ gR++k+c++rw +
MELO3C024799P1 9 KGPWSPEEDDALQRLVHKYGPRNWSLISKSIP-GRSGKSCRLRWCNQ 54
79******************************.***********985 PP
| |||||||
| 2 | Myb_DNA-binding | 54.1 | 3.5e-17 | 63 | 105 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
++T+eEde +++a +++G++ W+tIar + gRt++ +k++w++
MELO3C024799P1 63 AFTPEEDETIINAQALYGNK-WATIARLLS-GRTDNAIKNHWNST 105
79******************.*********.***********986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 26.114 | 4 | 59 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.08E-31 | 6 | 102 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.4E-17 | 8 | 57 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.0E-19 | 9 | 54 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.3E-26 | 10 | 62 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 3.96E-17 | 11 | 53 | No hit | No description |
| SMART | SM00717 | 2.7E-14 | 60 | 108 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 21.238 | 61 | 110 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.5E-22 | 63 | 109 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 4.9E-14 | 63 | 105 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 5.49E-12 | 63 | 106 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 285 aa Download sequence Send to blast |
MAKQIDRIKG PWSPEEDDAL QRLVHKYGPR NWSLISKSIP GRSGKSCRLR WCNQLSPQVE 60 HRAFTPEEDE TIINAQALYG NKWATIARLL SGRTDNAIKN HWNSTLKRKC SSMADESGGC 120 DATSPPLKRS LSAGLYMPPN SPSASDVSDS GFFPAVSSSH VYRPVARTGA VLPPGETVSS 180 SNDPPTSLSL SLPGADSSEV NFVANSAQGM GGVSERRREK EESNSNGFGI FGSDLMTVMQ 240 EMIRKEVRNY MAGLMEQNVG GGGGVCYQQA AAGGFRNVVI QRID* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 3e-39 | 6 | 112 | 4 | 110 | B-MYB |
| 1mse_C | 3e-39 | 8 | 110 | 3 | 105 | C-Myb DNA-Binding Domain |
| 1msf_C | 3e-39 | 8 | 110 | 3 | 105 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that functions in salt stress response. Acts as negative regulator of NHX7/SOS1 and CBL4/SOS3 induction in response to salt stress (PubMed:23809151). In response to auxin, activates the transcription of the auxin-responsive gene IAA19. The IAA19 transcription activation by MYB73 is enhanced by direct interaction between MYB73 and PYL8 (PubMed:24894996). {ECO:0000269|PubMed:23809151, ECO:0000269|PubMed:24894996}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00476 | DAP | Transfer from AT4G37260 | Download |
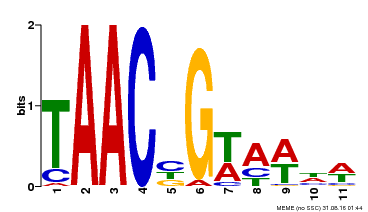
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by salt stress. {ECO:0000269|PubMed:23809151}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681915 | 0.0 | LN681915.1 Cucumis melo genomic scaffold, anchoredscaffold00070. | |||
| GenBank | LN713265 | 0.0 | LN713265.1 Cucumis melo genomic chromosome, chr_11. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008462845.1 | 0.0 | PREDICTED: transcription factor MYB44-like | ||||
| Swissprot | O23160 | 3e-91 | MYB73_ARATH; Transcription factor MYB73 | ||||
| TrEMBL | A0A1S3CID0 | 0.0 | A0A1S3CID0_CUCME; transcription factor MYB44-like | ||||
| STRING | XP_008462845.1 | 0.0 | (Cucumis melo) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF843 | 34 | 124 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G37260.1 | 5e-92 | myb domain protein 73 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



