 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MLOC_10556.5 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Hordeinae; Hordeum
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 787aa MW: 86776.1 Da PI: 5.1801 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 58.1 | 2e-18 | 50 | 96 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT eEd++l +av+ + g++Wk+Ia++++ gRt+ qc +rwqk+l
MLOC_10556.5 50 KGNWTREEDDILSRAVQTYNGKHWKKIAECFP-GRTDVQCLHRWQKVL 96
79******************************.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 61.5 | 1.7e-19 | 102 | 148 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+W++eEd+++v+ vk++G++ W+tIa+ ++ gR +kqc++rw+++l
MLOC_10556.5 102 KGPWSKEEDDIIVEMVKKYGPKKWSTIAQALP-GRIGKQCRERWHNHL 148
79******************************.*************97 PP
| |||||||
| 3 | Myb_DNA-binding | 49.4 | 1e-15 | 154 | 196 | 1 | 45 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwq 45
+++WT+eE+ l++a++++G++ W+ + ++ gRt++++k++w+
MLOC_10556.5 154 KDAWTQEEEITLIHAHRMYGNK-WAELTKFLP-GRTDNSIKNHWN 196
679*******************.*********.***********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 16.76 | 45 | 96 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 3.3E-16 | 46 | 103 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.1E-16 | 49 | 98 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.0E-15 | 50 | 96 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 7.6E-25 | 52 | 113 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 2.84E-14 | 52 | 96 | No hit | No description |
| PROSITE profile | PS51294 | 32.215 | 97 | 152 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.98E-31 | 99 | 195 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.2E-18 | 101 | 150 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.29E-16 | 104 | 148 | No hit | No description |
| Pfam | PF13921 | 2.9E-18 | 105 | 165 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.2E-26 | 114 | 155 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.9E-14 | 153 | 201 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 19.934 | 153 | 203 | IPR017930 | Myb domain |
| CDD | cd00167 | 3.00E-11 | 156 | 196 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.2E-21 | 156 | 203 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003713 | Molecular Function | transcription coactivator activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 787 aa Download sequence Send to blast |
MTSDKGKASK KAGEASGQPS TTHEGKVNNE PQRQRSINGR TTGPTRRSTK GNWTREEDDI 60 LSRAVQTYNG KHWKKIAECF PGRTDVQCLH RWQKVLNPEL IKGPWSKEED DIIVEMVKKY 120 GPKKWSTIAQ ALPGRIGKQC RERWHNHLNP GINKDAWTQE EEITLIHAHR MYGNKWAELT 180 KFLPGRTDNS IKNHWNSSVK KKIDSYMSSG LLAQVSRLPL VEHHAHFSSS PAITQQNSED 240 SESNAVREVE DSSGCSQSSL GIVSCSQVQD TNLTLSCDLH VNADPSKTEA HDSQSSMCQE 300 GYTSTEGVAS VMSEVHCHVP SSRFDPDTLL QQEISQRMNS QMDVDEIPGN SMFENNQTIC 360 STSNNERPML QYEIASDMPI SVLTNVSGAE QKLHFMSEAD FSSPNCLKSE LWQDISFQSL 420 LSGPDVVDAD SLSRPNHQSE THSSEANINF LAAPNPSHTS DPSSMMVVAY GQDPSMSVPQ 480 SLISNGLSDA ADEESREMQA SGSEMITCIH DSFGDSEQFA TPGSTDGRHG SSATIERIPE 540 YGDKQLTDAE EPAACMAKEP SMAQGEAVPD EKQDKGALFY EPPRFPSMDV PFVSCDLVTS 600 GDLQEYSPLG IRQLMRSTMS VTTPLRLWGS PTHDESPDVV LKSAAKSFIS TPSILKKRPR 660 DLLSPTSEKG IEKKSRTEQD SSVLGTSSVS AQTSCMHAIK DKAIVTESVF CTNRSSSFKP 720 LEKKLEFCDE NKDNLGESEQ AKDGRNAQNN YSVDEHARGE QCSTANMVNI NDEVSTSKSV 780 LQLHLPS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 1e-67 | 50 | 203 | 6 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 1e-67 | 50 | 203 | 6 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00466 | DAP | Transfer from AT4G32730 | Download |
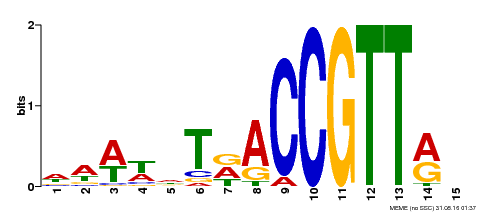
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | MLOC_10556.5 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HE996827 | 0.0 | HE996827.1 Triticum aestivum cv. Arina SNP, chromosome 3B, clone Taes_arina_ctg_69800. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020153262.1 | 0.0 | transcription factor MYB3R-1-like isoform X1 | ||||
| TrEMBL | M0UFS2 | 0.0 | M0UFS2_HORVV; Uncharacterized protein | ||||
| STRING | MLOC_10556.1 | 0.0 | (Hordeum vulgare) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G32730.1 | 1e-150 | Homeodomain-like protein | ||||



