 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MLOC_14401.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Hordeinae; Hordeum
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 415aa MW: 44990.3 Da PI: 6.2629 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 63 | 5.8e-20 | 109 | 154 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
rg+WT+eEdell+++v q+G + W+tI+ +++ gR +kqc++rw ++
MLOC_14401.1 109 RGPWTEEEDELLKRLVDQHGEHKWATISDHLP-GRIGKQCRERWTNH 154
89******************************.***********997 PP
| |||||||
| 2 | Myb_DNA-binding | 53.7 | 4.7e-17 | 165 | 204 | 4 | 45 |
S-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 4 WTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwq 45
WT+ +d ll+da+k +G++ W++Iar+++ gR+++ +k++w+
MLOC_14401.1 165 WTEADDILLIDAHKIHGNR-WSSIARCLP-GRSENAVKNHWN 204
*******************.*********.***********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 27.692 | 104 | 159 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.2E-18 | 108 | 157 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 2.55E-30 | 108 | 203 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 2.8E-18 | 109 | 154 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.1E-23 | 109 | 157 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 6.15E-17 | 111 | 155 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.2E-17 | 158 | 208 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 16.6 | 160 | 211 | IPR017930 | Myb domain |
| SMART | SM00717 | 9.3E-15 | 161 | 209 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.9E-14 | 165 | 204 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.11E-13 | 165 | 207 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010439 | Biological Process | regulation of glucosinolate biosynthetic process | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:1904095 | Biological Process | negative regulation of endosperm development | ||||
| GO:2000692 | Biological Process | negative regulation of seed maturation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 415 aa Download sequence Send to blast |
MASMGELLLN QELAAVAAMS TDAYGRFERY TLSPAHGFPH FQQAGSAPTV FPDTGLLVSG 60 FGMPPSTFIV MPEGTHAADA YGAEAVPVEI PVVRRQRPAA RNAGPTPFRG PWTEEEDELL 120 KRLVDQHGEH KWATISDHLP GRIGKQCRER WTNHVRPGIK KEHIWTEADD ILLIDAHKIH 180 GNRWSSIARC LPGRSENAVK NHWNATKRSL KSKRRFKKKT SQQAAPGQFT LLEEYIRDKM 240 MADENAAPPC PSSGGVGYDG QVVPGAAAML AVSGPPGMGQ YLHPADAAGS SSQAGVMNLS 300 APLPDLNAYG GEMLEGYYYP ATFPTYSDNN LLHHGPETAF PQMFSAQGSM HMHAACTNLN 360 LFPLPQHLGG GYYGSETGRS SAGGSSDQDE DVVQMASREF QTSEDEATLD LTGFS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 7e-39 | 105 | 212 | 3 | 110 | B-MYB |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00385 | DAP | Transfer from AT3G27785 | Download |
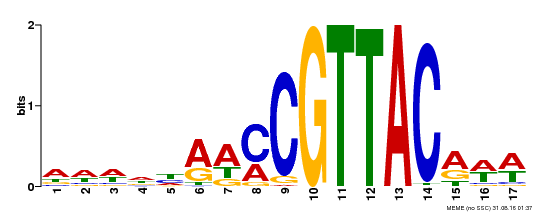
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | MLOC_14401.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG670306 | 0.0 | HG670306.1 Triticum aestivum chromosome 3B, genomic scaffold, cultivar Chinese Spring. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020163979.1 | 0.0 | uncharacterized protein LOC109749424 | ||||
| TrEMBL | M0UUP1 | 0.0 | M0UUP1_HORVV; Uncharacterized protein | ||||
| STRING | MLOC_14401.1 | 0.0 | (Hordeum vulgare) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1882 | 33 | 101 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G27785.1 | 3e-39 | myb domain protein 118 | ||||



