 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MLOC_18401.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Hordeinae; Hordeum
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 768aa MW: 86133.4 Da PI: 6.722 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 73.6 | 2.3e-23 | 76 | 177 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT.......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsef 90
f+k+lt sd++++g +++ +++a+e+ +++++ ++l+ +d++g +W++++i+r++++r++l++GW+ Fv++++L +gD ++F + +++
MLOC_18401.2 76 FCKTLTASDTSTHGGFSVLRRHADEClppldmtQSPPT--QELVAKDLHGMEWRFRHIFRGQPRRHLLQSGWSVFVSSKRLVAGDAFIFL--RGESG 168
99*****************************7555544..58************************************************..44999 PP
..EEEEE-S CS
B3 91 elvvkvfrk 99
el+v+v+r+
MLOC_18401.2 169 ELRVGVRRA 177
9*****996 PP
| |||||||
| 2 | Auxin_resp | 106.5 | 3.1e-35 | 202 | 283 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalkvkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
a+ha++tk++F+v+Y+Pr+s+seF+++++k+ +++k+ +s+G+Rfkm+fe+e+++e+r++Gt+vg +ld+ Wp+S WrsLk
MLOC_18401.2 202 AWHAINTKTMFTVYYKPRTSRSEFIIPYDKYTESVKNIYSIGTRFKMRFEGEEAPEQRFTGTIVGSDNLDQ-LWPESSWRSLK 283
89******************************************************************999.9********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 1.14E-38 | 69 | 205 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 7.9E-40 | 70 | 191 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 9.77E-21 | 74 | 176 | No hit | No description |
| Pfam | PF02362 | 3.1E-21 | 76 | 177 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 11.843 | 76 | 178 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 4.2E-23 | 76 | 178 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 5.9E-32 | 202 | 283 | IPR010525 | Auxin response factor |
| Pfam | PF02309 | 2.7E-5 | 622 | 674 | IPR033389 | AUX/IAA domain |
| SuperFamily | SSF54277 | 7.06E-14 | 632 | 715 | No hit | No description |
| PROSITE profile | PS51745 | 26.438 | 638 | 731 | IPR000270 | PB1 domain |
| Gene3D | G3DSA:3.10.20.240 | 6.3E-4 | 656 | 713 | No hit | No description |
| Pfam | PF02309 | 2.5E-8 | 679 | 723 | IPR033389 | AUX/IAA domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008285 | Biological Process | negative regulation of cell proliferation | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009911 | Biological Process | positive regulation of flower development | ||||
| GO:0010047 | Biological Process | fruit dehiscence | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0010227 | Biological Process | floral organ abscission | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0048481 | Biological Process | plant ovule development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 768 aa Download sequence Send to blast |
MNQVAANQMR LYDLPSKLLC RVLNVELKAE ADTDEVYAQV MLMPEPEQSE AATTTTEKSS 60 SATGGTMPAR PAVRSFCKTL TASDTSTHGG FSVLRRHADE CLPPLDMTQS PPTQELVAKD 120 LHGMEWRFRH IFRGQPRRHL LQSGWSVFVS SKRLVAGDAF IFLRGESGEL RVGVRRAMRQ 180 LSNIASSVIS SHSMHLGVLA TAWHAINTKT MFTVYYKPRT SRSEFIIPYD KYTESVKNIY 240 SIGTRFKMRF EGEEAPEQRF TGTIVGSDNL DQLWPESSWR SLKVRWDESS TIPRPDRVSP 300 WEIEPASSPP VNPLPLSRAK RSRPNVPPAS PESSVRTKEG ATKADMDCAQ AQRNQNNTVL 360 PGHEQRSNKL TDINDFDATV QKPMVWSTPP PNIGKNNPLT FQQRPSVHNS IQLRRREADF 420 KDASSGAQHF GDSLGFFMQT TFDEAPNRLG SFKNQFQDHS YARQFADPYL FMHQQPSLTV 480 ESSRKMHTEN NELHFWNGPG TVYGNSIDHV QDFRFKEHPS NWLSPQFSRA EQPRVIRPHA 540 SIAPIELEKT TEGSDFKIFG FKVDTASAGF NHLNSPMAAT HEPVLQTQPS VSLDHLQTDC 600 SPEVSLSIAG TTDNEKNIQQ CPQSSKDVQS KSHGASTRSC TKVHKQGVAL GRSVDLSKFV 660 DYDELTAELD KMFDFDGELM SSNKNWQIVY TDNEGDMMLV GDDPWEEFCS MVRKICIYTK 720 EEVQKMNSKP SDARKEEGSV EGDGATEKAH LPVSSDLNSS QLGCVKAD |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldv_A | 1e-142 | 1 | 308 | 56 | 357 | Auxin response factor 1 |
| 4ldw_A | 1e-142 | 1 | 308 | 56 | 357 | Auxin response factor 1 |
| 4ldw_B | 1e-142 | 1 | 308 | 56 | 357 | Auxin response factor 1 |
| 4ldx_A | 1e-142 | 1 | 308 | 56 | 357 | Auxin response factor 1 |
| 4ldx_B | 1e-142 | 1 | 308 | 56 | 357 | Auxin response factor 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). | |||||
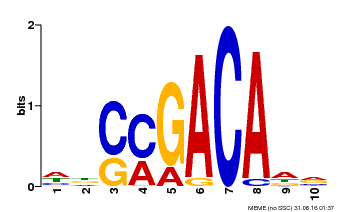
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00574 | DAP | Transfer from AT5G62000 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | MLOC_18401.2 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK372798 | 0.0 | AK372798.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv3013C04. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020197968.1 | 0.0 | auxin response factor 4-like | ||||
| Swissprot | Q5JK20 | 0.0 | ARFD_ORYSJ; Auxin response factor 4 | ||||
| TrEMBL | M0V4Z0 | 0.0 | M0V4Z0_HORVV; Auxin response factor | ||||
| STRING | MLOC_18401.1 | 0.0 | (Hordeum vulgare) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G62000.3 | 0.0 | auxin response factor 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



