 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MLOC_22013.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Hordeinae; Hordeum
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 154aa MW: 17422.6 Da PI: 10.1211 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 30.4 | 8.4e-10 | 1 | 46 | 11 | 56 |
HHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 11 qkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkev 56
++NR +A +sR+RKk +++ Le+k+k Leae ++L l+ e
MLOC_22013.2 1 MRNRDSAMKSRERKKSYVKDLETKSKYLEAECRRLSYALQCCAAEN 46
7*******************************99997777655555 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd14704 | 6.04E-11 | 1 | 47 | No hit | No description |
| Pfam | PF00170 | 8.0E-8 | 1 | 52 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 3.2E-11 | 1 | 54 | No hit | No description |
| SuperFamily | SSF57959 | 1.38E-9 | 1 | 53 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0030968 | Biological Process | endoplasmic reticulum unfolded protein response | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005789 | Cellular Component | endoplasmic reticulum membrane | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 154 aa Download sequence Send to blast |
MRNRDSAMKS RERKKSYVKD LETKSKYLEA ECRRLSYALQ CCAAENMALR QNMLKDRPIG 60 AHTVMQESAV LMETLPLVSL LCLVSIVCLF LTPGLPNRSL VAPRRAERDL AMVAGKPSSD 120 QPETLELLLH GRRWRGTRER IKLDILPLRA AAAC |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in endoplasmic reticulum (ER) stress response (PubMed:21223397, PubMed:22050533, PubMed:22199238). Acts downstream of the ER stress sensors IRE1, BZIP39 and BZIP60 to activate BiP chaperone genes (PubMed:22050533, PubMed:22199238). {ECO:0000269|PubMed:21223397, ECO:0000269|PubMed:22050533, ECO:0000269|PubMed:22199238}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00019 | PBM | Transfer from AT1G42990 | Download |
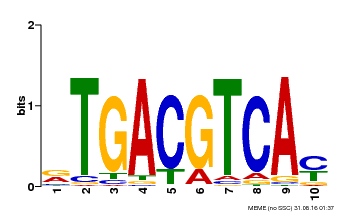
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | MLOC_22013.2 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By dithiothreitol-induced endoplasmic reticulum (ER) stress response (PubMed:22050533, PubMed:21223397). Induced by salt stress (PubMed:22050533). {ECO:0000269|PubMed:21223397, ECO:0000269|PubMed:22050533}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK365505 | 0.0 | AK365505.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv2034K24. | |||
| GenBank | AK368565 | 0.0 | AK368565.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, partial cds, clone: NIASHv2076C04. | |||
| GenBank | AK369957 | 0.0 | AK369957.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv2101P04. | |||
| GenBank | AK370149 | 0.0 | AK370149.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv2105L10. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020184631.1 | 1e-103 | bZIP transcription factor 50 | ||||
| Swissprot | Q69XV0 | 1e-73 | BZP50_ORYSJ; bZIP transcription factor 50 | ||||
| TrEMBL | A0A287XD86 | 1e-107 | A0A287XD86_HORVV; Uncharacterized protein | ||||
| TrEMBL | A0A287XDD5 | 1e-107 | A0A287XDD5_HORVV; Uncharacterized protein | ||||
| STRING | MLOC_22013.1 | 1e-108 | (Hordeum vulgare) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G42990.1 | 4e-14 | basic region/leucine zipper motif 60 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



