 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MLOC_3107.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Hordeinae; Hordeum
|
||||||||
| Family | GATA | ||||||||
| Protein Properties | Length: 247aa MW: 26059.3 Da PI: 8.9754 | ||||||||
| Description | GATA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GATA | 52.8 | 5.4e-17 | 158 | 195 | 1 | 36 |
GATA 1 CsnCgttk..TplWRrgpdgnktLCnaCGlyyrkkglk 36
C++Cg ++ Tp++Rrgpdg++tLCnaCGl ++ kg++
MLOC_3107.2 158 CQHCGISSnnTPMMRRGPDGPRTLCNACGLAWANKGMM 195
*****99889*************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00979 | 5.9E-8 | 28 | 63 | IPR010399 | Tify domain |
| PROSITE profile | PS51320 | 13.136 | 28 | 63 | IPR010399 | Tify domain |
| Pfam | PF06200 | 1.4E-10 | 30 | 62 | IPR010399 | Tify domain |
| Pfam | PF06203 | 2.3E-17 | 88 | 130 | IPR010402 | CCT domain |
| PROSITE profile | PS51017 | 13.404 | 88 | 130 | IPR010402 | CCT domain |
| SMART | SM00401 | 2.0E-12 | 152 | 205 | IPR000679 | Zinc finger, GATA-type |
| SuperFamily | SSF57716 | 1.24E-11 | 155 | 199 | No hit | No description |
| Gene3D | G3DSA:3.30.50.10 | 3.2E-15 | 157 | 198 | IPR013088 | Zinc finger, NHR/GATA-type |
| CDD | cd00202 | 9.75E-14 | 157 | 196 | No hit | No description |
| Pfam | PF00320 | 2.9E-14 | 158 | 193 | IPR000679 | Zinc finger, GATA-type |
| PROSITE pattern | PS00344 | 0 | 158 | 185 | IPR000679 | Zinc finger, GATA-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 247 aa Download sequence Send to blast |
MDEEAAAAAA HAHHGAAGPL GMEAGGAPQA PSNTLTLSFQ GEVFVFESVS AEKVQAVLLL 60 LGGRELGPGM GAGPSSSASY SKRLNSHRMA SLMRFREKRK ERNFDKKIRY SVRKEVAHRM 120 HRHKGQFTSS KAKAEEAASV ANSDWGAVEG RPPSAPVCQH CGISSNNTPM MRRGPDGPRT 180 LCNACGLAWA NKGMMREVKG HTPLKVVPPA TDDAQNGVAE APTAEQQHLA LEAAPEAPAP 240 AANGHDS |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that specifically binds 5'-GATA-3' or 5'-GAT-3' motifs within gene promoters. {ECO:0000250|UniProtKB:Q8LAU9}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00369 | DAP | Transfer from AT3G21175 | Download |
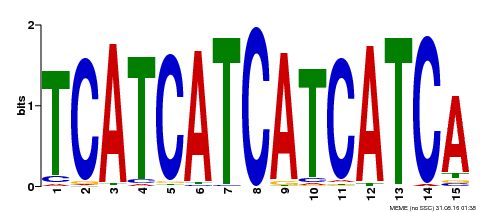
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | MLOC_3107.2 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By drought and salt stresses. {ECO:0000269|PubMed:19618278}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK358896 | 0.0 | AK358896.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv1085G16. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020166796.1 | 1e-132 | GATA transcription factor 17-like isoform X1 | ||||
| Swissprot | Q6Z433 | 1e-104 | GAT17_ORYSJ; GATA transcription factor 17 | ||||
| TrEMBL | A0A287TK54 | 1e-180 | A0A287TK54_HORVV; Uncharacterized protein | ||||
| STRING | MLOC_3107.1 | 0.0 | (Hordeum vulgare) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G21175.2 | 1e-53 | ZIM-like 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



