 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MLOC_326.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Hordeinae; Hordeum
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 277aa MW: 29415.4 Da PI: 10.4967 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 130.8 | 4.9e-41 | 146 | 222 | 1 | 77 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S-- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkq 77
+Cq+egC+adls ak+yhrrhkvCe h+ka+ v+++g++qrfCqqCsrfh l+efDe+krsCr+rLa+hn+rrrk++
MLOC_326.1 146 RCQAEGCKADLSGAKHYHRRHKVCEYHAKASLVAANGKQQRFCQQCSRFHVLTEFDEAKRSCRKRLAEHNRRRRKPA 222
6**************************************************************************86 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 4.9E-35 | 139 | 208 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.479 | 144 | 221 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 5.76E-39 | 145 | 224 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 3.1E-31 | 147 | 220 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009554 | Biological Process | megasporogenesis | ||||
| GO:0009556 | Biological Process | microsporogenesis | ||||
| GO:0048653 | Biological Process | anther development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 277 aa Download sequence Send to blast |
MVQAGGGGGG GQRHLQGAMM YDNLDFAAAF GQFQDAPHHQ MLGMPPNGSG AGGLVPMAPP 60 PMPMPGMQLQ MPPMAMHGHG DVYPTLGMVK REGGGAGDAG RIGLNLGRRT YFSPGDMMAV 120 DRLLMRSRLG GVFGLGFGGA HHQPPRCQAE GCKADLSGAK HYHRRHKVCE YHAKASLVAA 180 NGKQQRFCQQ CSRFHVLTEF DEAKRSCRKR LAEHNRRRRK PASSSTTATS KDSAPPSKKP 240 NPGAISGSSA ADNNTLSATK STISSNTSAI SCLQQHN |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 2e-30 | 147 | 220 | 11 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 203 | 220 | KRSCRKRLAEHNRRRRKP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
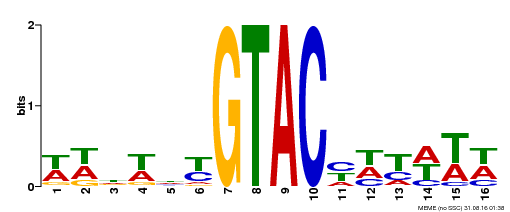
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00090 | SELEX | Transfer from AT1G02065 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | MLOC_326.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KF447886 | 0.0 | KF447886.1 Triticum aestivum squamosa promoter-binding-like protein 22 (SPL22) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020180500.1 | 1e-143 | squamosa promoter-binding-like protein 10 | ||||
| Swissprot | A2YFT9 | 2e-82 | SPL10_ORYSI; Squamosa promoter-binding-like protein 10 | ||||
| Swissprot | Q0DAE8 | 2e-82 | SPL10_ORYSJ; Squamosa promoter-binding-like protein 10 | ||||
| TrEMBL | A0A3B6SN70 | 1e-152 | A0A3B6SN70_WHEAT; Uncharacterized protein | ||||
| STRING | MLOC_326.1 | 0.0 | (Hordeum vulgare) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2375 | 35 | 93 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G02065.1 | 8e-44 | squamosa promoter binding protein-like 8 | ||||



