 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MLOC_43537.4 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Hordeinae; Hordeum
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 249aa MW: 26357.2 Da PI: 4.1792 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 76.2 | 4.2e-24 | 181 | 230 | 1 | 51 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQ 51
k++++WtpeLH+rFv+aveqL G++kA+P++ile+m++++Lt+++++SHLQ
MLOC_43537.4 181 KAKVDWTPELHRRFVQAVEQL-GIDKAVPSRILEIMGINSLTRHNIASHLQ 230
6799*****************.***************************** PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 13.3 | 178 | 237 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.1E-23 | 179 | 230 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 2.51E-15 | 179 | 231 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.2E-23 | 181 | 231 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.0E-6 | 184 | 230 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007165 | Biological Process | signal transduction | ||||
| GO:0009658 | Biological Process | chloroplast organization | ||||
| GO:0009910 | Biological Process | negative regulation of flower development | ||||
| GO:0010380 | Biological Process | regulation of chlorophyll biosynthetic process | ||||
| GO:0010638 | Biological Process | positive regulation of organelle organization | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:1900056 | Biological Process | negative regulation of leaf senescence | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 249 aa Download sequence Send to blast |
MLAVSSARCL AADADEQRAE AAPMETVGGA AVVADLDIDF DFTVDDIDFG DFFLRLEDGD 60 ALPDLEVDPA DIFTDLEAAA AGVQELQDQQ VPCAFLAAVE DVGSVSSAGG VVGVENTAFG 120 EEGRLGDEKR GCNQAEVGEN MSGGDRPIVP DAKSPSSTTS SSTEAESRHK SSGKSSHGKK 180 KAKVDWTPEL HRRFVQAVEQ LGIDKAVPSR ILEIMGINSL TRHNIASHLQ VRAGMELETN 240 VKLNIDCSV |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 5e-15 | 178 | 230 | 2 | 54 | ARR10-B |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcriptional activator that promotes chloroplast development. Acts as an activator of nuclear photosynthetic genes involved in chlorophyll biosynthesis, light harvesting, and electron transport. {ECO:0000269|PubMed:19808806}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00022 | PBM | Transfer from AT2G20570 | Download |
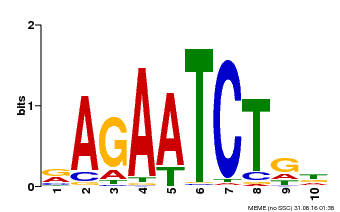
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | MLOC_43537.4 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By light. {ECO:0000269|PubMed:11340194}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK361623 | 0.0 | AK361623.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv1145D04. | |||
| GenBank | AK367495 | 0.0 | AK367495.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv2058D05. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020191527.1 | 1e-140 | probable transcription factor GLK1 | ||||
| Swissprot | Q5Z5I4 | 2e-62 | GLK1_ORYSJ; Probable transcription factor GLK1 | ||||
| TrEMBL | F2DCK6 | 1e-164 | F2DCK6_HORVV; Predicted protein | ||||
| STRING | MLOC_43537.1 | 1e-165 | (Hordeum vulgare) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G44190.1 | 3e-24 | GOLDEN2-like 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



