 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MLOC_43575.3 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Hordeinae; Hordeum
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 367aa MW: 41139.8 Da PI: 9.3255 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 54.8 | 2.3e-17 | 41 | 90 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++grW+++I+d + k+++lg f+ta Aa+a+++a+ k++g
MLOC_43575.3 41 SQYRGVTFYRRTGRWESHIWD------CgKQVYLGGFDTAHAAARAYDRAAIKFRG 90
78*******************......55************************998 PP
| |||||||
| 2 | AP2 | 41.9 | 2.4e-13 | 133 | 183 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpseng...krkrfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV+ +k grW+A+ g +k+++lg f+++ eAa+a+++a+ +++g
MLOC_43575.3 133 SKYRGVTLHK-CGRWEAR---M---GqllGKKYIYLGLFDSEVEAARAYDRAAIRFNG 183
89********.7******...5...334336**********99**********99987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 4.18E-16 | 41 | 99 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 1.5E-9 | 41 | 90 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 7.77E-11 | 41 | 100 | No hit | No description |
| Gene3D | G3DSA:3.30.730.10 | 2.5E-17 | 42 | 99 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 17.622 | 42 | 98 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.0E-31 | 42 | 104 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 3.44E-25 | 133 | 193 | No hit | No description |
| Pfam | PF00847 | 7.3E-9 | 133 | 183 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 2.81E-17 | 133 | 192 | IPR016177 | DNA-binding domain |
| Gene3D | G3DSA:3.30.730.10 | 2.8E-16 | 134 | 192 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 16.318 | 134 | 191 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 2.4E-33 | 134 | 197 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.2E-6 | 135 | 146 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.2E-6 | 173 | 193 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 367 aa Download sequence Send to blast |
QAPAPPPTAP VWQPRRAEEL VVAQRVAPKK KTRRGPRSRS SQYRGVTFYR RTGRWESHIW 60 DCGKQVYLGG FDTAHAAARA YDRAAIKFRG LEADINFNLS DYEEDLKQMR NWTKEEFVHI 120 LRRQSTGFAR GSSKYRGVTL HKCGRWEARM GQLLGKKYIY LGLFDSEVEA ARAYDRAAIR 180 FNGRDAVTNF DSSSYNGDAT PDVENEAIVD ADALDLDLRM SQPTAHDPKR DNIIAGLQLT 240 FDSPESSTTM VSSQPMSSSS QWPVHQHGTA VPPQQHQRLY PSACHGFYPN VQVQVQERPL 300 EPRPPEPSSF PGWGWHAQAV PPGSSHSLLL YAAASSGFST AAGANPAPPP SYPDHHHRFY 360 FPRPPDN |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor (By similarity). Involved in spikelet transition (Probable). Together with SNB, controls synergistically inflorescence architecture and floral meristem establishment via the regulation of spatio-temporal expression of B- and E-function floral organ identity genes in the lodicules and of spikelet meristem genes (PubMed:22003982). Prevents lemma and palea elongation as well as grain growth (PubMed:28066457). {ECO:0000250|UniProtKB:P47927, ECO:0000269|PubMed:22003982, ECO:0000269|PubMed:28066457, ECO:0000305|PubMed:26631749}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00615 | PBM | Transfer from AT2G28550 | Download |
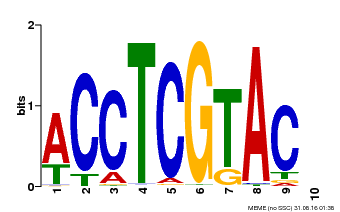
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | MLOC_43575.3 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Target of miR172 microRNA mediated cleavage, particularly during floral organ development. {ECO:0000269|PubMed:22003982, ECO:0000305|PubMed:20017947, ECO:0000305|PubMed:26631749, ECO:0000305|PubMed:28066457}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK355002 | 0.0 | AK355002.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv1014I14. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020154436.1 | 0.0 | AP2-like ethylene-responsive transcription factor TOE3 isoform X2 | ||||
| Swissprot | Q84TB5 | 1e-154 | AP22_ORYSJ; APETALA2-like protein 2 | ||||
| TrEMBL | F2CTQ0 | 0.0 | F2CTQ0_HORVV; Predicted protein | ||||
| STRING | MLOC_43575.3 | 0.0 | (Hordeum vulgare) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2448 | 38 | 90 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G36920.2 | 2e-89 | AP2 family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



