 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MLOC_43575.6 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Hordeinae; Hordeum
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 255aa MW: 28263.3 Da PI: 7.1684 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 37.8 | 4.9e-12 | 25 | 69 | 1 | 49 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpseng...krkrfslgkfgtaeeAakaaiaa 49
s+y+GV+ +k grW+A+ g +k+++lg f+++ eAa+aai++
MLOC_43575.6 25 SKYRGVTLHK-CGRWEAR---M---GqllGKKYIYLGLFDSEVEAARAAIRF 69
89********.7******...5...334336**********99*******97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 6.2E-8 | 25 | 69 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 6.31E-21 | 25 | 81 | No hit | No description |
| SuperFamily | SSF54171 | 3.79E-12 | 25 | 80 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 10.652 | 26 | 79 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 7.4E-25 | 26 | 85 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 8.9E-10 | 26 | 80 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 255 aa Download sequence Send to blast |
MRNWTKEEFV HILRRQSTGF ARGSSKYRGV TLHKCGRWEA RMGQLLGKKY IYLGLFDSEV 60 EAARAAIRFN GRDAVTNFDS SSYNGDATPD VENEAIVDAD ALDLDLRMSQ PTAHDPKRDN 120 IIAGLQLTFD SPESSTTMVS SQPMSSSSQW PVHQHGTAVP PQQHQRLYPS ACHGFYPNVQ 180 VQVQERPLEP RPPEPSSFPG WGWHAQAVPP GSSHSLLLYA AASSGFSTAA GANPAPPPSY 240 PDHHHRFYFP RPPDN |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor (By similarity). Involved in spikelet transition (Probable). Together with SNB, controls synergistically inflorescence architecture and floral meristem establishment via the regulation of spatio-temporal expression of B- and E-function floral organ identity genes in the lodicules and of spikelet meristem genes (PubMed:22003982). Prevents lemma and palea elongation as well as grain growth (PubMed:28066457). {ECO:0000250|UniProtKB:P47927, ECO:0000269|PubMed:22003982, ECO:0000269|PubMed:28066457, ECO:0000305|PubMed:26631749}. | |||||
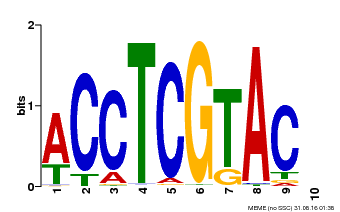
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00615 | PBM | Transfer from AT2G28550 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | MLOC_43575.6 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Target of miR172 microRNA mediated cleavage, particularly during floral organ development. {ECO:0000269|PubMed:22003982, ECO:0000305|PubMed:20017947, ECO:0000305|PubMed:26631749, ECO:0000305|PubMed:28066457}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK355002 | 0.0 | AK355002.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv1014I14. | |||
| GenBank | AK356946 | 0.0 | AK356946.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv1042M02. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020154436.1 | 1e-138 | AP2-like ethylene-responsive transcription factor TOE3 isoform X2 | ||||
| Swissprot | Q84TB5 | 7e-93 | AP22_ORYSJ; APETALA2-like protein 2 | ||||
| TrEMBL | A0A023R8L7 | 1e-161 | A0A023R8L7_TRITU; Spelt factor protein (Fragment) | ||||
| TrEMBL | K4NRN1 | 1e-161 | K4NRN1_TRIDC; Spelt factor protein (Fragment) | ||||
| STRING | MLOC_43575.3 | 1e-160 | (Hordeum vulgare) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G28550.3 | 5e-44 | related to AP2.7 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



