 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | MLOC_52321.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Hordeinae; Hordeum
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 328aa MW: 35327.2 Da PI: 7.4382 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 49.2 | 1.4e-15 | 41 | 72 | 47 | 78 |
SSEEETTT--SS--S-STTTT-------S--- CS
SBP 47 srfhelsefDeekrsCrrrLakhnerrrkkqa 78
rfh l efDe krsCr+rL++hn+rrrk+q+
MLOC_52321.1 41 CRFHLLGEFDEVKRSCRKRLDGHNRRRRKPQP 72
69***************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51141 | 13.015 | 1 | 70 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 3.92E-12 | 41 | 74 | IPR004333 | Transcription factor, SBP-box |
| Gene3D | G3DSA:4.10.1100.10 | 4.7E-4 | 41 | 57 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 1.5E-7 | 42 | 69 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0048653 | Biological Process | anther development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 328 aa Download sequence Send to blast |
MTGALFFFVL ASDQDLYSSF HAEFVVVYLE KCLLPDQTVV CRFHLLGEFD EVKRSCRKRL 60 DGHNRRRRKP QPDPLNPAGL FANHHGVPRF ASYPQIFSPT SMAEPKWPGG IAVKTEADAF 120 HEQYYSFNGA ASLFHHGKPE RKHFPFLTDG GDATFGCQPP AFTITPSSES SSNSSRHSNG 180 KTTMFAHDGG PDHNCALSLL SDNPAPAHIM NPAEAHHLGG GGGVTIQYGG GGGGKVARLH 240 NNGDVSLTGL SYVSLGDKGA PMLLHTSNRS QHAAATATTA VTTSTAPAPT ASQMQQYHGY 300 YHHQHQVGAD QGNHQDAGGM HALPFSSW |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 52 | 69 | KRSCRKRLDGHNRRRRKP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' (By similarity). May be involved in panicle development. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00555 | DAP | Transfer from AT5G50570 | Download |
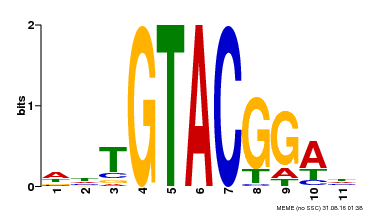
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | MLOC_52321.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Negatively regulated by microRNAs miR156b and miR156h. {ECO:0000305|PubMed:16861571}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK356077 | 0.0 | AK356077.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv1029L02. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020178580.1 | 1e-166 | squamosa promoter-binding-like protein 2 | ||||
| Swissprot | Q0JGI1 | 1e-66 | SPL2_ORYSJ; Squamosa promoter-binding-like protein 2 | ||||
| TrEMBL | M0WKB0 | 0.0 | M0WKB0_HORVV; Uncharacterized protein | ||||
| STRING | MLOC_52321.2 | 0.0 | (Hordeum vulgare) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G50670.1 | 2e-16 | SBP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



